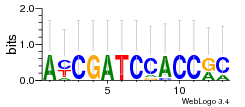
Module 19 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.000000000021 | 0.0000048 | 82 | 25 | 20160204 |
| MSMEG_2974 Peptidyl-prolyl cis-trans isomerase B PpiB (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase, cyclophilin-type) | MSMEG_1548 Glycerol dehydratase (EC 4.2.1.30) (Propanediol utilization: dehydratase, medium subunit) |
| MSMEG_2975 Metallo-beta-lactamase family protein (Putative glyoxalase II (Hydroxyacylglutathione hydrolase)) (EC 3.1.2.6) | MSMEG_5720 Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35) (Putative 3-hydroxyacyl-CoA dehydrogenase) |
| MSMEG_2976 Histidine--tRNA ligase (EC 6.1.1.21) (Histidyl-tRNA synthetase) | MSMEG_5721 Acetyl-CoA acetyltransferase (Acetyl-CoA acyltransferase/thiolase) (EC 2.3.1.16) |
| MSMEG_3055 S-adenosylmethionine synthase (AdoMet synthase) (EC 2.5.1.6) (MAT) (Methionine adenosyltransferase) | MSMEG_1545 Uncharacterized protein |
| MSMEG_4501 Sodium:dicarboxylate symporter (Sodium:dicarboxylate symporter: DAACS family) | MSMEG_1546 Coenzyme B12-dependent glycerol dehydrogenase small subunit |
| MSMEG_3784 Transcriptional regulator IclR (Transcriptional regulator, IclR family protein) | MSMEG_1547 Glycerol dehydratase large subunit (EC 4.2.1.30) (Uncharacterized protein) |
| MSMEG_3782 Endoribonuclease L-PSP (Endoribonuclease L-PSP, putative) | MSMEG_6322 Diacylglycerol O-acyltransferase (EC 2.3.1.20) |
| MSMEG_2351 Electron transfer flavoprotein beta subunit (Electron transfer flavoprotein, beta subunit) | MSMEG_3744 Tyrosine recombinase XerD |
| MSMEG_2352 Electron transfer flavoprotein, alpha subunit | MSMEG_3745 MutT/nudix family protein (NTP pyrophosphohydrolase including oxidative damage repair enzymes) (EC 3.6.1.13) |
| MSMEG_1346 50S ribosomal protein L11 | MSMEG_3746 CTP synthase (EC 6.3.4.2) (CTP synthetase) (UTP--ammonia ligase) |
| MSMEG_1347 50S ribosomal protein L1 | MSMEG_6586 Alpha/beta fold hydrolase (EC 3.-.-.-) (Alpha/beta hydrolase, putative) |
| MSMEG_1344 Preprotein translocase secE1 (Translocase) | MSMEG_6822 Beta-lactamase (Putative conserved alanine rich lipoprotein LppW) |
| MSMEG_1345 Transcription termination/antitermination protein NusG |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.4.of.7..ethambutol.treatment
X2XR.rep1
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.3.of.6..isoniazid.treatment
Wild.type.rep2.y
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
Sub.MIC.INH
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
Slow.vs..Fast.Replicate.2
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
X4XR.rep1
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
dosR.WT_Replicate3
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments