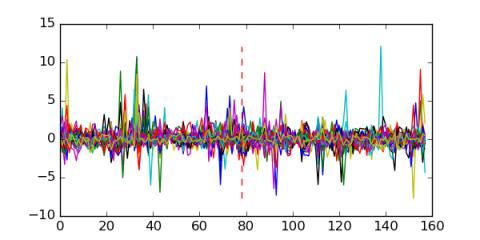
Module 31 Residual: 0.63
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.63 | 0.0000000000031 | 0.00000065 | 78 | 34 | 20160204 |
| MSMEG_1598 Uncharacterized protein | MSMEG_1486 RNA polymerase sigma-70 factor |
| MSMEG_3338 Oxidoreductase, FAD/FMN-binding | MSMEG_4004 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100) |
| MSMEG_4030 Putative membrane spanning protein (Uncharacterized protein) | MSMEG_4005 Calcium-binding protein (SMP-30/Gluconolaconase/LRE domain protein) (EC 3.1.1.17) |
| MSMEG_3551 Linalool 8-monooxygenase (EC 1.14.13.151) | MSMEG_3797 Putative esterase family protein |
| MSMEG_0518 sn-glycerol-3-phosphate import ATP-binding protein UgpC (EC 3.6.3.20) | MSMEG_5450 Redox-sensitive transcriptional activator SoxR |
| MSMEG_5596 Oxidoreductase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_4698 Uncharacterized protein |
| MSMEG_5595 MarR family transcriptional regulator (MarR-family protein transcriptional regulator) | MSMEG_4697 Uncharacterized protein |
| MSMEG_5448 Arginine deiminase (ADI) (EC 3.5.3.6) (Arginine dihydrolase) | MSMEG_3128 Uncharacterized protein |
| MSMEG_1285 SEC-C motif domain protein (Tetratricopeptide repeat family protein) | MSMEG_2682 Transcriptional regulator |
| MSMEG_1001 Acetyltransferase, GNAT family (Acetyltransferase, gnat family protein, putative) | MSMEG_2680 Amino acid transporter |
| MSMEG_6925 Uncharacterized protein | MSMEG_2681 Proline imino-peptidase |
| MSMEG_4363 Uncharacterized protein | MSMEG_6048 Cobalamin synthesis protein P47K (Cobalamin synthesis protein/P47K) |
| MSMEG_4365 Uncharacterized protein | MSMEG_6045 ABC heavy metal transporter, inner membrane subunit (ABC-3) |
| MSMEG_4029 Uncharacterized protein | MSMEG_6046 ABC transporter related protein (Cation ABC transporter, ATP-binding protein, putative) |
| MSMEG_3328 Uncharacterized protein | MSMEG_6047 Cation ABC transporter, periplasmic cation-binding protein, putative |
| MSMEG_6662 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) | MSMEG_2150 |
| MSMEG_1487 Putative transmembrane anti-sigma factor (Uncharacterized protein) | MSMEG_5064 Malyl-CoA lyase |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Sub.MIC.INH
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
WT_log.phase_rep2
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
X4XR.rep2
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.4
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.2.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
dosR.WT_Replicate3
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments