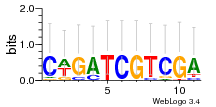
Module 58 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.00000000087 | 110 | 79 | 30 | 20160204 |
| MSMEG_3643 Uncharacterized protein | MSMEG_5212 Uncharacterized protein |
| MSMEG_6614 Integral membrane protein (Metallophosphoesterase) | MSMEG_4655 AraC-family transcriptional regulator (Transcriptional regulator, AraC family protein) |
| MSMEG_4444 Uncharacterized protein | MSMEG_4654 Aldo/keto reductase (EC 1.1.1.91) |
| MSMEG_1491 Histidinol-phosphate aminotransferase 2 (EC 2.6.1.9) | MSMEG_4745 ErfK/YbiS/YcfS/YnhG family protein |
| MSMEG_4644 Probable molybdenum cofactor guanylyltransferase (MoCo guanylyltransferase) (EC 2.7.7.77) (GTP:molybdopterin guanylyltransferase) (Mo-MPT guanylyltransferase) (Molybdopterin guanylyltransferase) (Molybdopterin-guanine dinucleotide synthase) | MSMEG_4670 Glycosyltransferase, MGT family (EC 2.4.1.17) (N-glycosylation) |
| MSMEG_6355 Uncharacterized protein | MSMEG_2008 Uncharacterized protein |
| MSMEG_0599 Acyl-CoA synthase | MSMEG_6005 Uncharacterized protein |
| MSMEG_4976 Isochorismatase hydrolase | MSMEG_6684 Agarase (EC 3.2.1.81) |
| MSMEG_4975 Flavin-nucleotide-binding protein (Pyridoxamine 5'-phosphate oxidase-related, FMN-binding protein) | MSMEG_1963 Putative transcriptional regulatory protein |
| MSMEG_2538 MarR-family protein transcriptional regulator | MSMEG_6225 Major facilitator superfamily MFS_1 (Proton antiporter efflux pump) |
| MSMEG_0597 Uncharacterized protein | MSMEG_5787 Uncharacterized protein |
| MSMEG_0019 Amino acid adenylation (Peptide synthetase) | MSMEG_1355 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_0035 FHA domain protein | MSMEG_1134 Protease HtpX homolog (EC 3.4.24.-) |
| MSMEG_1306 Aldehyde dehydrogenase (NAD) family protein (Aldehyde dehydrogenase (NAD+)) (EC 1.2.1.3) | MSMEG_1132 FAD binding domain, putative (Putative oxidoreductase) |
| MSMEG_4454 L-threonine aldolase, low-specificity (EC 4.1.2.5) | MSMEG_1176 PE-PPE, C-terminal domain protein (Uncharacterized protein) |
Biological.replicate.4.of.7..falcarinol.treatment
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Biological.replicate.2.of.6..isoniazid.treatment
X2XR.rep1
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Wild.type.rep1.x
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.2.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.1.of.6..negative.control
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
Sub.MIC.INH
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
stphcy5plogcy3slide580norm
X3dbfcy5plogcy3slide601norm
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
WT_stationary.phase_rep2
WT_stationary.phase_rep1
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.6..kanamycin.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
WT.vs..DglnR.N_starvation_rep1
X4dbfcy3plogcy5slide903norm
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Comments
Log in to post comments