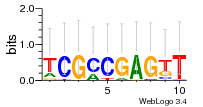
Module 76 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.00000000086 | 0.013 | 76 | 36 | 20160204 |
| MSMEG_3706 Isocitrate lyase (EC 4.1.3.1) (Putative isocitrate lyase aceab second part (Isocitrase) (Isocitratase)) | MSMEG_2984 Putative hydrolase (Uncharacterized protein) |
| MSMEG_6489 2,6-dihydroxypyridine hydroxylase (Monooxygenase FAD-binding protein) (EC 1.14.13.1) | MSMEG_3254 RDD domain containing protein (RDD family protein, putative) |
| MSMEG_3052 Uncharacterized protein | MSMEG_3255 DoxX (DoxX subfamily protein, putative) |
| MSMEG_5735 Alpha-keto-acid decarboxylase (KDC) (EC 4.1.1.-) | MSMEG_0363 TetR-family protein regulatory protein |
| MSMEG_5734 Alpha-methylacyl-CoA racemase (Fatty-acid-CoA racemase Far) (EC 5.1.99.4) | MSMEG_0362 Amidohydrolase 2 |
| MSMEG_1289 Transcriptional regulator, XRE family (Uncharacterized protein) | MSMEG_5090 Uncharacterized protein |
| MSMEG_4401 Phosphonoacetaldehyde hydrolase | MSMEG_6251 Uncharacterized protein |
| MSMEG_0808 Chalcone synthase pks10 (EC 2.3.1.74) (Possible chalcone synthase Pks10) | MSMEG_0185 MmpL6 protein |
| MSMEG_3464 Alcohol dehydrogenase (Alcohol dehydrogenase (ADH-HT) oxidoreductase-like protein) (EC 1.1.1.1) | MSMEG_3215 ABC transporter, ATP-binding component (ABC-type molybdenum transport system, ATPase component/photorepair protein PhrA) |
| MSMEG_2253 Uncharacterized protein | MSMEG_0898 Ketose-bisphosphate aldolase (EC 4.1.2.13) (Tagatose-1,6-bisphosphate aldolase GatY) (EC 4.1.2.-) |
| MSMEG_6372 GtrA-like protein (Membrane protein) | MSMEG_1290 Dihydroxy-acid dehydratase (DAD) (EC 4.2.1.9) |
| MSMEG_4481 Uncharacterized protein | MSMEG_3257 Xylulokinase (EC 2.7.1.17) |
| MSMEG_4884 Uncharacterized protein | MSMEG_1306 Aldehyde dehydrogenase (NAD) family protein (Aldehyde dehydrogenase (NAD+)) (EC 1.2.1.3) |
| MSMEG_3612 Probable conserved integral membrane protein | MSMEG_2286 |
| MSMEG_4345 Uncharacterized protein | MSMEG_0268 Transcriptional regulator, GntR family protein |
| MSMEG_5746 Gas vesicle protein | MSMEG_2916 DNA-binding response regulator, PhoP family protein |
| MSMEG_0888 Phosphoesterase, PA-phosphatase related protein (Uncharacterized protein) | MSMEG_2917 Uncharacterized protein |
| MSMEG_5325 Uncharacterized protein | MSMEG_2915 Sensor histidine kinase (Sensor protein) (EC 2.7.13.3) |
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
X4XR.rep1
Biological.replicate.4.of.7..ethambutol.treatment
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
Biological.replicate.4.of.6..kanamycin.treatment
Wild.type.rep2.y
Wild.type.rep1.y
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
WT_log.phase_rep2
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.2.of.6..negative.control
X4dbfcy5plogcy3slide480norm
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments