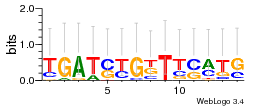
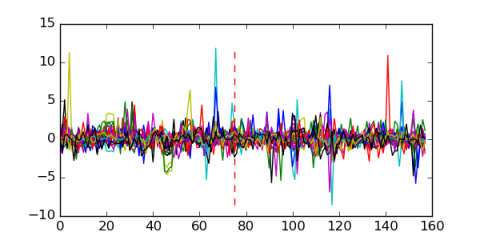
Module 99 Residual: 0.63
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.63 | 0.00000000034 | 0.23 | 75 | 28 | 20160204 |
| MSMEG_4503 Uncharacterized protein | MSMEG_4181 Phosphoribosyl-ATP pyrophosphatase (PRA-PH) (EC 3.6.1.31) |
| MSMEG_6730 Putative oxidoreductase YdbC (EC 1.-.-.-) | MSMEG_4180 ATP phosphoribosyltransferase (ATP-PRT) (ATP-PRTase) (EC 2.4.2.17) |
| MSMEG_5243 Helix-turn-helix motif (Pyridoxamine 5'-phosphate oxidase-related, FMN-binding protein) | MSMEG_1644 Uncharacterized protein |
| MSMEG_5242 Diacylglycerol O-acyltransferase (EC 2.3.1.20) | MSMEG_3906 tRNA (Adenine-N(1)-)-methyltransferase |
| MSMEG_4754 K+-dependent Na+/Ca+ exchanger related-protein (Na+/Ca+ antiporter, CaCA family) | MSMEG_2802 Uncharacterized protein |
| MSMEG_4755 Peptidase M20 | MSMEG_4434 Cof protein (Cof-like hydrolase) |
| MSMEG_3397 Acetylornithine deacetylase (Acetylornithine deacetylase or succinyl-diaminopimelate desuccinylase) (EC 3.5.1.16) | MSMEG_4381 Acetamidase/Formamidase (Amidase) |
| MSMEG_5344 Uncharacterized protein | MSMEG_4382 Dehydrogenase/reductase SDR family protein member 10 (EC 1.1.-.-) (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_5345 Glycosyl hydrolases family protein 16 | MSMEG_5255 Conserved hypothetical transmembrane protein (Uncharacterized protein) |
| MSMEG_1764 L-lysine-epsilon aminotransferase (EC 2.6.1.36) | MSMEG_6577 Methylmalonyl-CoA carboxyltransferase 12S subunit (EC 2.1.3.1) |
| MSMEG_0609 Helix-turn-helix domain protein (Transcriptional regulator, XRE family) | MSMEG_4499 Uncharacterized protein |
| MSMEG_0607 Methyltransferase, putative, family protein | MSMEG_6206 Serine/threonine protein kinase PksC (EC 2.7.11.1) (Serine/threonine-protein kinase) |
| MSMEG_0278 Uncharacterized protein | MSMEG_6207 Uncharacterized protein |
| MSMEG_0646 Phosphate-import permease protein PhnE | MSMEG_3621 Membrane NADH dehydrogenase NdhA (EC 1.6.99.3) (NADH dehydrogenase) |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep1
Bedaquiline.vs..DMSO.15min
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.7..panaxydol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
dcpA.knockout.rep2
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
DglnR_without.vs..with.N_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep2.x
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments