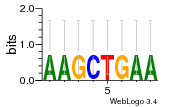
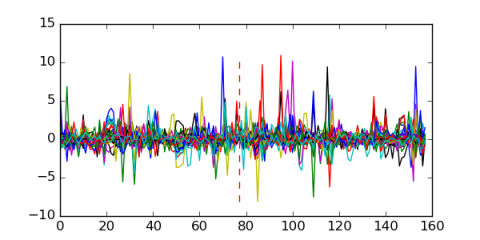
Module 107 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 14000 | 10000 | 77 | 32 | 20160204 |
| MSMEG_3529 Fatty acid hydroxylase | MSMEG_6600 Transposase |
| MSMEG_6841 Uncharacterized protein | MSMEG_1482 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_1482/MSMEI_1446 (EC 2.1.1.-) |
| MSMEG_6755 Isoflavone reductase | MSMEG_3573 Integral membrane protein (Integral membrane protein, putative) |
| MSMEG_4121 GntR family transcriptional regulator (GntR-family protein transcriptional regulator) | MSMEG_0424 Hsp20/alpha crystallin family protein |
| MSMEG_1806 Uncharacterized protein | MSMEG_1615 Gamma-glutamyltranspeptidase (EC 2.3.2.2) |
| MSMEG_0666 Acetyl-CoA synthetase, putative (EC 6.2.1.14) | MSMEG_6473 O-methyltransferase-like protein (Tetracenomycin polyketide synthesis O-methyltransferase TcmP) (EC 2.1.1.-) |
| MSMEG_1479 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_1479/MSMEI_1443 (EC 2.1.1.-) | MSMEG_3470 NAD-dependent epimerase/dehydratase (EC 5.1.3.2) |
| MSMEG_3802 Tetratricopeptide TPR_4 (Tetratricopeptide repeat family protein) | MSMEG_2203 Alpha/beta hydrolase |
| MSMEG_2005 Sugar phosphate isomerase/epimerase (Xylose isomerase-like TIM barrel) | MSMEG_1987 Uncharacterized protein |
| MSMEG_0234 Metallopeptidase (PgPepO oligopeptidase Metallo peptidase MEROPS family M13) (EC 3.4.24.-) | MSMEG_0103 Major facilitator family protein transporter (Major facilitator superfamily MFS_1) |
| MSMEG_5336 Amidate substrates transporter protein (Putative transporter protein amiS2) | MSMEG_0128 Putative thioesterase (Thioesterase superfamily protein) |
| MSMEG_1081 Uncharacterized protein | MSMEG_5025 AraC-family transcriptional regulator (Putative transcriptional regulator) |
| MSMEG_3290 Regulatory protein (Regulatory protein LuxR) | MSMEG_2448 Uncharacterized protein |
| MSMEG_1481 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_1481/MSMEI_1445 (EC 2.1.1.-) | MSMEG_2918 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) |
| MSMEG_6453 Sulfate permease | MSMEG_2919 2-dehydropantoate 2-reductase (Ketopantoate reductase ApbA/PanE-like protein) (EC 1.1.1.169) |
| MSMEG_1480 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_1480/MSMEI_1444 (EC 2.1.1.-) | MSMEG_3587 Amino acid permease-associated region (D-serine/D-alanine/glycine transporter) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Plogcy5plogcy3slide113norm
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Bedaquiline.vs..DMSO.30min
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
dosR.WT_Replicate2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
X4XR.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep1
Sub.MIC.INH
X3dbfcy5plogcy3slide601norm
X3dbfcy3plogcy5slide607norm
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep1
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments