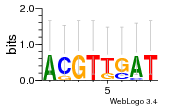
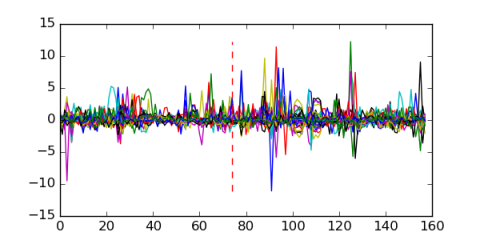
Module 146 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.000057 | 0.00037 | 74 | 30 | 20160204 |
| MSMEG_3138 Thioredoxin (Thioredoxin TrxB1) | MSMEG_2566 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase (EC 1.1.1.53) |
| MSMEG_1867 Transposase (Transposase IS3/IS911) | MSMEG_2565 4-hydroxy-2-oxovalerate aldolase (EC 4.1.2.-) (HpcH/HpaI aldolase) (EC 4.1.2.20) |
| MSMEG_3137 NADH:flavin oxidoreductase/NADH oxidase (Oxidoreductase) | MSMEG_2564 Alcohol dehydrogenase, zinc-binding protein (Oxidoreductase, zinc-binding dehydrogenase family protein) |
| MSMEG_1970 Sigma factor (Transcriptional regulator) (EC 2.7.13.3) | MSMEG_2563 Cytochrome P450 superfamily protein |
| MSMEG_5610 Putative transcriptional regulator family protein (Transcriptional regulator, HxlR family) | MSMEG_1812 Uncharacterized protein |
| MSMEG_5364 Amidohydrolase 2 | MSMEG_1813 Propionyl-CoA carboxylase beta chain (Propionyl-CoA carboxylase beta chain 5 AccD5) (EC 6.4.1.3) |
| MSMEG_4870 FAD binding domain, putative (Fumarate reductase/succinate dehydrogenase flavoprotein-like protein) | MSMEG_3369 Major facilitator superfamily MFS_1 (Putative transport protein) |
| MSMEG_0530 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) | MSMEG_2526 Amine oxidase (EC 1.4.3.-) |
| MSMEG_1811 Maf-like protein MSMEG_1811/MSMEI_1767 | MSMEG_2568 Alpha/beta hydrolase fold-3 (LipW protein) |
| MSMEG_5878 Cutinase (EC 3.1.1.74) | MSMEG_0280 Alpha/beta hydrolase fold (Alpha/beta hydrolase fold-3) |
| MSMEG_5609 Carotenoid oxygenase | MSMEG_0281 Choline dehydrogenase (Glucose-methanol-choline oxidoreductase) (EC 1.1.99.1) |
| MSMEG_5608 Acetyl-CoA acetyltransferase (Putative lipid carrier protein or keto acyl-coa thiolase) (EC 2.3.1.-) | MSMEG_0282 Putative ethyl tert-butyl ether degradation protein EthD (Uncharacterized protein) |
| MSMEG_6742 Uncharacterized protein | MSMEG_2930 Uncharacterized protein |
| MSMEG_5604 Integral membrane protein | MSMEG_2588 Cobyric acid synthase |
| MSMEG_2567 Cyclohexanone monooxygenase (EC 1.14.13.22) (Probable monooxygenase) (EC 1.14.13.-) | MSMEG_6829 Transcriptional regulator, HxlR family (Transcriptional regulatory protein) |
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
dcpA.knockout.rep2
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Wild.type.rep1.y
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
Slow.vs..Fast.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
X4dbfcy5plogcy3slide901norm
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..negative.control
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.2.of.7..ethambutol.treatment
dosR.WT_Replicate2
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
X2XR.rep1
X2XR.rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Bedaquiline.vs..DMSO..45min
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments