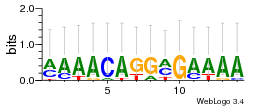
Module 150 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 1.5e-18 | 0.0000000027 | 81 | 23 | 20160204 |
| MSMEG_3159 Methylmalonyl-CoA mutase large subunit (Methylmalonyl-CoA mutase large subunit mutB) (EC 5.4.99.2) | MSMEG_5174 Regulatory protein GntR, HTH |
| MSMEG_5484 Uncharacterized protein | MSMEG_3160 LAO/AO transport system ATPase (EC 2.7.-.-) |
| MSMEG_3157 Integral membrane protein (Uncharacterized protein) | MSMEG_4200 Peptidase M20 |
| MSMEG_5636 Metal cation transporter ATPase P-type ctpE (P-type ATPase-metal cation transport) | MSMEG_1193 TROVE domain protein |
| MSMEG_5637 Penicillin-binding protein 4 | MSMEG_4041 Hydroxyquinol 1,2-dioxygenase (EC 1.13.11.-) (EC 1.13.11.37) |
| MSMEG_3445 5,10-methylenetetrahydromethanopterin reductase (EC 1.5.99.11) | MSMEG_0082 Conserved membrane protein (Uncharacterized protein) |
| MSMEG_3444 Choline dehydrogenase (Glucose-methanol-choline oxidoreductase) (EC 1.1.99.1) | MSMEG_0083 Membrane-anchored mycosin mycp1 (Peptidase S8 and S53, subtilisin, kexin, sedolisin) |
| MSMEG_4669 Protein FdhD homolog | MSMEG_3585 MmpS2 protein |
| MSMEG_3158 Methylmalonyl-CoA mutase, beta subunit (EC 5.4.99.2) (Methylmalonyl-CoA mutase, small subunit) | MSMEG_4727 Mycocerosic acid synthase (EC 2.3.1.111) |
| MSMEG_1764 L-lysine-epsilon aminotransferase (EC 2.6.1.36) | MSMEG_0123 Uncharacterized protein |
| MSMEG_6311 Saccharopine dehydrogenase (EC 1.5.1.7) | MSMEG_0122 Putative periplasmic solute-binding protein (Uncharacterized protein UPF0065) |
| MSMEG_5175 NAD-dependent protein deacylase Sir2 (EC 3.5.1.-) (Regulatory protein SIR2 homolog) |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.3.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
X4XR.rep1
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
Wild.type.rep2.y
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.3.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
Biological.replicate.4.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
Biological.replicate.7.of.7..panaxydol.treatment
stphcy3plogcy5slide581norm
Biological.replicate.3.of.6..negative.control
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
mc2155.vs.Dcrp1.replicate.1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
Biological.replicate.1.of.7..panaxydol.treatment
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate1
X4dbfcy5plogcy3slide901norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments