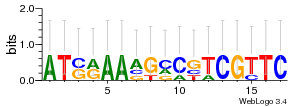
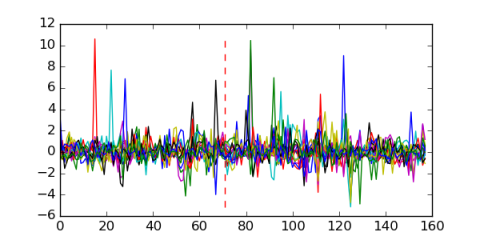
Module 174 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 160 | 9500 | 71 | 16 | 20160204 |
| MSMEG_6860 Uncharacterized protein | MSMEG_0036 Uncharacterized protein |
| MSMEG_0671 Alcohol dehydrogenase GroES domain protein (S-(Hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) | MSMEG_4735 Uncharacterized protein |
| MSMEG_1226 Sulfatase-modifying factor 1 | MSMEG_4419 Glucose-1-dehydrogenase |
| MSMEG_0423 Uncharacterized protein | MSMEG_4050 Uncharacterized protein |
| MSMEG_0979 Keratin associated protein | MSMEG_3881 Proline dipeptidase (Putative dipeptidase pepE) (EC 3.4.13.9) |
| MSMEG_3351 Uncharacterized protein | MSMEG_0720 Integral membrane protein |
| MSMEG_0576 Conserved transmembrane transport protein MmpL1 (MmpL4 protein) | MSMEG_2477 Cyclopentanol dehydrogenase (EC 1.1.1.163) |
| MSMEG_1587 Alpha/beta hydrolase fold (Alpha/beta hydrolase fold protein) | MSMEG_0050 Uncharacterized protein |
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.5.of.7..ethambutol.treatment
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
stphcy3plogcy5slide581norm
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.2.of.6..kanamycin.treatment
Biological.replicate.6.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
mc2155.vs.Dcrp1.replicate.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
dosR.WT_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Biological.replicate.4.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
X2XR.rep1
X2XR.rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments