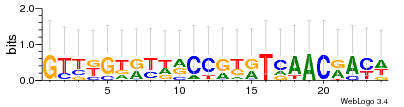
Module 188 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 3.6e-21 | 0.0000013 | 77 | 21 | 20160204 |
| MSMEG_0974 Cytochrome c-type biogenesis protein CcsB | MSMEG_0972 Cytochrome C biogenesis protein transmembrane region (Cytochrome C-type biogenesis protein ccdA) |
| MSMEG_1743 Fatty acid desaturase | MSMEG_0973 Conserved membrane protein |
| MSMEG_1741 TetR-family protein transcriptional regulator | MSMEG_0970 Phosphoglycerate mutase (EC 5.4.2.-) (Phosphoglycerate mutase family protein) |
| MSMEG_0086 1-phosphofructokinase (EC 2.7.1.56) (Fructose-1-phosphate kinase and related fructose-6-phosphate kinase (PfkB) (1-phosphofructokinase protein)) (EC 2.7.1.56) | MSMEG_0904 Probable conserved membrane protein (Uncharacterized protein) |
| MSMEG_6702 6-oxohexanoate dehydrogenase, succinate-semialdehyde dehydrogenase (NADP+) (EC 1.2.1.16) ([NADP+] succinate-semialdehyde dehydrogenase) | MSMEG_1742 Oxidoreductase (Oxidoreductase FAD-binding region) |
| MSMEG_6701 Amino acid permease (Amino acid permease-associated region) | MSMEG_3246 Response regulator (Two-component system response regulator) |
| MSMEG_6700 Regulatory protein (Transcriptional regulator, GntR family) | MSMEG_0969 Glutamate-1-semialdehyde 2,1-aminomutase (GSA) (EC 5.4.3.8) (Glutamate-1-semialdehyde aminotransferase) (GSA-AT) |
| MSMEG_0903 Dihydrolipoyl dehydrogenase (EC 1.8.1.4) | MSMEG_0971 Thiol-disulfide isomerase/thioredoxin (Uncharacterized protein) |
| MSMEG_0902 Cyclopropane-fatty-acyl-phospholipid synthase 1 (Mycolic acid synthase) (EC 2.1.1.79) | MSMEG_0084 Phosphocarrier protein HPr (EC 2.7.1.69) (Phosphocarrier protein hpr) |
| MSMEG_0088 Phosphoenolpyruvate-protein phosphotransferase (EC 2.7.3.9) (Phosphotransferase system, enzyme I) | MSMEG_0087 DeoR family transcriptional regulator (Glucitol operon repressor) |
| MSMEG_0085 PTS system, Fru family protein, IIABC components (Putative PTS fructose-specific enzyme IIABC) (EC 2.7.1.69) |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.4.of.6..negative.control
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep1
DglnR_without.vs..with.N_rep1
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.2.of.7..ethambutol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.2.of.7..falcarinol.treatment
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT.vs..DamtR.N.surplus_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
Biological.replicate.2.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments