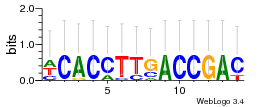
Module 218 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.00014 | 0.0000065 | 79 | 32 | 20160204 |
| MSMEG_5045 D-2-hydroxyglutarate dehydrogenase (EC 1.1.99.-) (Putative glyoclate oxidase FAD-linked subunit) | MSMEG_5193 Transcriptional regulator, AraC family protein |
| MSMEG_5044 ATPase (Uncharacterized protein) | MSMEG_3695 Uncharacterized protein |
| MSMEG_6442 Integral membrane protein | MSMEG_5194 Integral membrane protein |
| MSMEG_3676 Phenoxybenzoate dioxygenase beta subunit (EC 1.-.-.-) (Putative Iron-sulfur oxidoreductase) (EC 1.14.12.7) | MSMEG_4062 Glutamyl-tRNA(Gln) amidotransferase subunit A (EC 6.3.5.-) |
| MSMEG_0296 Transcriptional regulator, MarR family (Transcriptional regulator, MarR family protein) | MSMEG_0446 Amino acid permease-associated region (Putrescine importer) |
| MSMEG_3241 Uncharacterized protein | MSMEG_0445 Amidohydrolase family protein (Uncharacterized protein) |
| MSMEG_3116 Inositol-1-monophosphatase (Inositol-phosphate phosphatase) (EC 3.1.3.25) | MSMEG_6183 Membrane-associated serine protease (Serine protease) |
| MSMEG_5956 NAD dependent epimerase/dehydratase family protein (UDP-glucose 4-epimerase, GalE5) | MSMEG_6185 NTP pyrophosphohydrolase (NUDIX hydrolase) |
| MSMEG_6332 Amino acid ABC transporter, permease protein (Binding-protein-dependent transport systems inner membrane component) | MSMEG_6186 Putative membrane-anchored thioredoxin-like protein (Thiol-disulfide interchange related protein) (Uncharacterized protein) |
| MSMEG_6333 Amino acid ABC transporter, ATP-binding protein (Osmoprotectant (Glycine betaine/carnitine/choline/L-proline) transport ATP-binding protein ABC transporter proV) (EC 3.6.3.32) | MSMEG_3627 Urease subunit gamma (EC 3.5.1.5) (Urea amidohydrolase subunit gamma) |
| MSMEG_6331 ABC transporter, permease protein ProZ (Binding-protein-dependent transport systems inner membrane component) | MSMEG_3626 Urease subunit beta (EC 3.5.1.5) (Urea amidohydrolase subunit beta) |
| MSMEG_0035 FHA domain protein | MSMEG_3625 Urease subunit alpha (EC 3.5.1.5) (Urea amidohydrolase subunit alpha) |
| MSMEG_2960 Preprotein translocase, YajC subunit (Protein translocase subunit yajC) | MSMEG_3624 Urease accessory protein ureF (Urease accessory protein uref) |
| MSMEG_6857 Putative transcription regulator (Transcriptional regulator, IclR family) | MSMEG_3623 Urease accessory protein UreG |
| MSMEG_3675 Metal-dependent hydrolase (Uncharacterized protein) | MSMEG_3622 Uncharacterized protein |
| MSMEG_1890 3-phosphoshikimate 1-carboxyvinyltransferase (EC 2.5.1.19) | MSMEG_6806 Excinuclease ABC (Excinuclease ABC, A subunit) |
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
dosR.WT_Replicate4
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
DglnR_without.vs..with.N_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
Wild.type.rep3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Biological.replicate.1.of.7..falcarinol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_rep1
WT.with.vs..without.N_rep1
X2XR.rep1
Biological.replicate.5.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
dosR.WT_Replicate2
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments