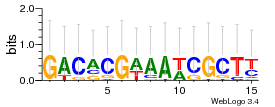
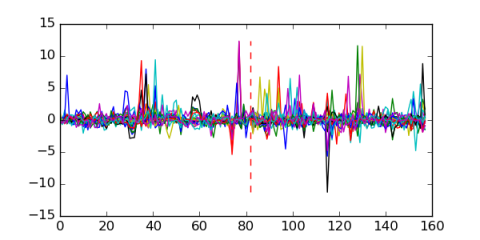
Module 232 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 4.2e-36 | 0.00034 | 82 | 26 | 20160204 |
| MSMEG_2235 Amidohydrolase 2 (Amidohydrolase family protein) | MSMEG_6417 Uncharacterized protein |
| MSMEG_6019 ABC-type sugar transport system ATPase component (Putative sugar ABC transporter ATP-binding protein) (EC 3.6.3.17) | MSMEG_4806 Acyl-CoA dehydrogenase (Putative acyl-CoA dehydrogenase) |
| MSMEG_6018 Xylose transport system permease protein XylH (EC 3.6.3.17) | MSMEG_4805 Acyl-CoA dehydrogenase |
| MSMEG_2149 Uncharacterized protein | MSMEG_3348 Bacterial extracellular solute-binding protein, family protein 7 (TRAP dicarboxylate transporter DctP subunit) |
| MSMEG_5971 Uncharacterized protein | MSMEG_6022 Xylose repressor, ROK-family protein transcriptional regulator |
| MSMEG_3292 DNA-binding protein (Transcriptional regulator, XRE family) | MSMEG_2244 Rieske (2Fe-2S) region (Rieske [2Fe-2S] domain protein) |
| MSMEG_3291 Uncharacterized protein | MSMEG_6020 D-xylose-binding periplasmic protein (EC 3.6.3.17) (Periplasmic binding protein/LacI transcriptional regulator) |
| MSMEG_4433 Dehydrogenase | MSMEG_6021 Xylose isomerase (EC 5.3.1.5) |
| MSMEG_4431 Putative regulatory protein (Uncharacterized protein) | MSMEG_2245 Bile-acid 7-alpha dehydratase (EC 4.2.1.106) (Uncharacterized protein) |
| MSMEG_4104 Major facilitator superfamily MFS_1 (Transporter, major facilitator family protein) | MSMEG_2079 Alcohol dehydrogenase (Alcohol dehydrogenase, zinc-binding protein) |
| MSMEG_1962 Uncharacterized protein | MSMEG_1961 Uncharacterized protein |
| MSMEG_1960 Uncharacterized protein | MSMEG_2813 Sensor kinase |
| MSMEG_4049 Sugar transporter, permease protein | MSMEG_2812 C-5 sterol desaturase (EC 1.3.-.-) (Membrane-bound C-5 Sterol desaturase erg3) |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
dcpA.knockout.rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.3.of.7..falcarinol.treatment
X2XR.rep1
dosR.WT_Replicate2
KO_log.phase_rep2
KO_log.phase_rep1
DglnR_without.vs..with.N_rep1
Wild.type.rep1.y
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Sub.MIC.INH
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.2.of.7..falcarinol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
X3dbfcy5plogcy3slide601norm
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..kanamycin.treatment
WT_stationary.phase_rep1
X4XR.rep1
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.with.vs..without.N_rep2
Biological.replicate.4.of.7..ethambutol.treatment
WT.with.vs..without.N_rep1
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
Biological.replicate.2.of.6..negative.control
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
WT.vs..DglnR.N_starvation_dye.swap_rep1
Comments
Log in to post comments