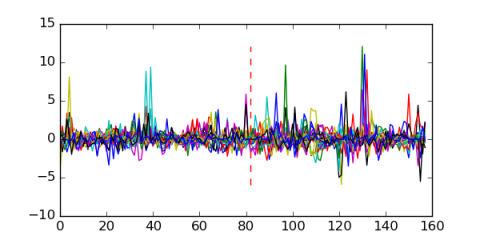
Module 254 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 3.1e-23 | 0.000000017 | 82 | 22 | 20160204 |
| MSMEG_3442 Cyclohexadienyl dehydratase (EC 4.2.1.-) (EC 4.2.1.51) (EC 4.2.1.91) | MSMEG_1924 GCN5-related N-acetyltransferase (N-acetyltransferase Ats1) (EC 2.3.1.-) |
| MSMEG_3441 Uncharacterized protein | MSMEG_1925 Isochorismate synthase DhbC (EC 5.4.4.2) |
| MSMEG_3440 L-serine ammonia-lyase (EC 4.3.1.17) | MSMEG_0733 Dihydrodipicolinate reductase (EC 1.17.1.8) (Dihydrodipicolinate reductase, N-terminus domain protein) |
| MSMEG_6929 Integral membrane protein MviN (Integral membrane protein MviN, putative) | MSMEG_0734 Rieske (2Fe-2S) region (Rieske [2Fe-2S] domain protein) |
| MSMEG_6211 Uncharacterized protein | MSMEG_0735 Putative transcriptional regulator |
| MSMEG_6212 Hemerythrin HHE cation binding domain subfamily protein, putative (Hemerythrin HHE cation binding region) | MSMEG_3438 Uncharacterized protein |
| MSMEG_2474 Cutinase (EC 3.1.1.74) | MSMEG_5027 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) (EC 4.4.1.5) |
| MSMEG_6928 Uncharacterized protein | MSMEG_5025 AraC-family transcriptional regulator (Putative transcriptional regulator) |
| MSMEG_6927 MutT/nudix family protein | MSMEG_4889 General substrate transporter (Integral membrane transport protein) |
| MSMEG_6926 Poly(A) polymerase PcnA (EC 2.7.7.-) (tRNA adenylyltransferase) (EC 2.7.7.72) | MSMEG_4888 Periplasmic binding proteins and sugar binding domain of the LacI family protein, putative (Transcriptional regulator, LacI family) |
| MSMEG_1926 Phosphoglycerate mutase family protein (Putative phosphoglycerate mutase gpm2) (EC 5.4.2.-) | MSMEG_5028 HIT family protein (Histidine triad (HIT) protein) (EC 3.6.1.17) |
KO_stationary.phase_rep1
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
X4XR.rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.5.of.7..ethambutol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
KO_log.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Wild.type.rep1.y
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT_log.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
dosR.WT_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Plogcy5plogcy3slide113norm
Biological.replicate.1.of.6..kanamycin.treatment
X3dbfcy3plogcy5slide607norm
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
stphcy5plogcy3slide487norm
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.5.of.6..isoniazid.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep1
Wild.type.rep2.x
Biological.replicate.1.of.6..negative.control
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
Biological.replicate.2.of.6..negative.control
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments