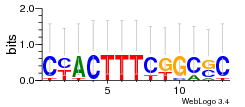
Module 314 Residual: 0.60
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.60 | 0.0000007 | 0.00025 | 79 | 27 | 20160204 |
| MSMEG_3263 Xylulose kinase | MSMEG_6413 Serine--tRNA ligase (EC 6.1.1.11) (Seryl-tRNA synthetase) (SerRS) (Seryl-tRNA(Ser/Sec) synthetase) |
| MSMEG_5571 ABC transporter ATP-binding protein (Sugar ABC transporter ATP-binding protein) | MSMEG_6412 Uncharacterized protein |
| MSMEG_5573 Sugar ABC transporter permease protein | MSMEG_5541 Glucose-6-phosphate isomerase (GPI) (EC 5.3.1.9) (Phosphoglucose isomerase) (PGI) (Phosphohexose isomerase) (PHI) |
| MSMEG_5572 Sugar ABC transporter permease protein | MSMEG_6415 Uncharacterized protein |
| MSMEG_5575 Repressor | MSMEG_6414 Uncharacterized protein |
| MSMEG_5574 Substrate binding protein | MSMEG_3234 Integral membrane protein |
| MSMEG_5577 Fructokinase (Fructokinase, PfkB) (EC 2.7.1.4) | MSMEG_3235 ABC-type amino acid transport system, secreted component (Extracellular solute-binding protein family 3) |
| MSMEG_5576 D-mannonate oxidoreductase (EC 1.1.1.57) | MSMEG_3236 ABC-type amino acid transport system, permease component (Truncated polar amino acid ABC transporter) |
| MSMEG_1249 ISMsm7, transposase orfB (Integrase catalytic region) | MSMEG_3237 ATP-binding protein |
| MSMEG_3094 Oxidoreductase, zinc-binding dehydrogenase family protein | MSMEG_1250 ISMsm7, transposase orfA (Transposase IS3/IS911) |
| MSMEG_3092 Transcriptional regulator, DeoR family (Transcriptional regulator, sugar-binding family protein) | MSMEG_0102 Putative acyl-CoA dehydrogenase |
| MSMEG_3093 Carbohydrate kinase FGGY (EC 2.7.1.16) (Putative sugar kinase protein) (EC 2.7.1.-) | MSMEG_0100 Phosphotyrosine protein phosphatase PtpB (EC 3.1.3.48) (Phosphotyrosine protein phosphatase ptpb) |
| MSMEG_4106 | MSMEG_4198 Dihydroorotate dehydrogenase (quinone) (EC 1.3.5.2) (DHOdehase) (DHOD) (DHODase) (Dihydroorotate oxidase) |
| MSMEG_4105 |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Biological.replicate.2.of.6..negative.control
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.2.of.7..panaxydol.treatment
DglnR_without.vs..with.N_rep1
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
MB100pruR.vs..MB100.rep.2
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
Wild.type.rep3
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.2.of.7..ethambutol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep1
Sub.MIC.INH
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.5.of.7..ethambutol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments