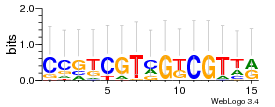
Module 362 Residual: 0.59
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.59 | 1.6e-20 | 0.000044 | 78 | 32 | 20160204 |
| MSMEG_5517 Conserved integral membrane protein (Uncharacterized protein) | MSMEG_1246 Uncharacterized protein |
| MSMEG_5516 Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (5'-phosphoribosylglycinamide transformylase) (GAR transformylase) | MSMEG_5935 ATP-dependent DNA helicase (ATP-dependent DNA helicase RecQ) |
| MSMEG_0862 Molybdopterin biosynthesis protein MoeA | MSMEG_5978 GlcNAc-PI de-N-acetylase family protein (Uncharacterized protein) |
| MSMEG_0863 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) | MSMEG_0250 Drug exporters of the RND superfamily-like protein (Membrane protein, MmpL family protein) |
| MSMEG_0864 Bacterial membrane flanked domain family protein | MSMEG_2801 Uncharacterized protein |
| MSMEG_5512 Magnesium chelatase | MSMEG_1922 3-dehydroquinate dehydratase (3-dehydroquinase) (EC 4.2.1.10) (Type II DHQase) |
| MSMEG_5511 von Willebrand factor type A (von Willebrand factor, type A) | MSMEG_1923 Membrane protein |
| MSMEG_5979 Putative glucose-1-phosphate cytidylyltransferase (EC 2.7.7.33) (Transferase) | MSMEG_1921 UPF0225 protein MSMEG_1921 |
| MSMEG_6617 NUDIX hydrolase (Nudix hydrolase) | MSMEG_0811 Monooxygenase, FAD-binding protein (Oxidoreductase) |
| MSMEG_5515 Bifunctional purine biosynthesis protein PurH | MSMEG_1586 Beta-lactamase |
| MSMEG_5513 Serine/threonine-protein kinase PknE (EC 2.7.11.1) | MSMEG_5980 Methyltransferase (NDP-hexose 3-C-methyltransferase protein) |
| MSMEG_2896 Uncharacterized protein | MSMEG_4924 Adenylate cyclase (EC 4.6.1.1) (Adenylate cyclase, family protein 3) |
| MSMEG_2895 Short chain oxidoreductase (Short-chain dehydrogenase/reductase SDR) | MSMEG_0346 MCE-family protein MCE3a (Virulence factor) |
| MSMEG_4050 Uncharacterized protein | MSMEG_1632 Uncharacterized protein |
| MSMEG_0809 Isoprenylcysteine carboxyl methyltransferase | MSMEG_1949 Uncharacterized protein |
| MSMEG_1588 Uncharacterized protein | MSMEG_2367 Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B (Asp/Glu-ADT subunit B) (EC 6.3.5.-) |
Biological.replicate.5.of.6..negative.control
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X4XR.rep1
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.5.of.6..isoniazid.treatment
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
Biological.replicate.4.of.6..negative.control
Biological.replicate.2.of.6..kanamycin.treatment
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
MB100pruR.vs..MB100.rep.2
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.1.of.6..kanamycin.treatment
X3dbfcy3plogcy5slide607norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT_log.phase_rep1
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Wild.type.rep1.x
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
dosR.WT_Replicate3
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
dosR.WT_Replicate2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments