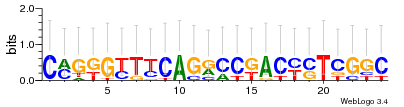
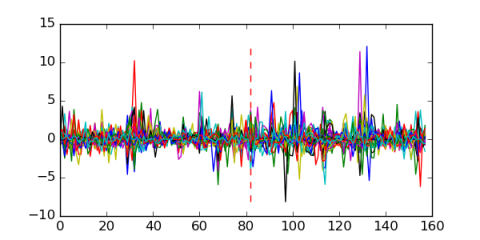
Module 377 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 0.00000000000011 | 0.00000000000019 | 82 | 32 | 20160204 |
| MSMEG_3050 Integration host factor (Integration host factor MihF) | MSMEG_2772 Amino acid permease (Putative conserved integral membrane alanine and valine and leucine rich protein (Amino acid permease, putative)) |
| MSMEG_3051 Guanylate kinase (EC 2.7.4.8) (GMP kinase) | MSMEG_2773 Conserved alanine valine and glycine rich protein (Putative RNA methyltransferase) (EC 2.1.1.-) |
| MSMEG_5613 Probable conserved transmembrane protein | MSMEG_2771 TrkA protein (TrkA-N) |
| MSMEG_6932 Uncharacterized protein | MSMEG_5364 Amidohydrolase 2 |
| MSMEG_1643 Uncharacterized protein | MSMEG_4416 Uncharacterized protein |
| MSMEG_1049 Methyltransferase type 11 (Methyltransferase, UbiE/COQ5 family protein) | MSMEG_6289 Trypsin |
| MSMEG_2083 Inositol monophosphatase (Putative histidinol-phosphate phosphatase, inositol monophosphatase family) (EC 3.1.3.15) | MSMEG_1777 Uncharacterized protein (UsfY protein) |
| MSMEG_4815 Thioesterase (Uncharacterized protein) | MSMEG_1776 Ribonuclease Z (RNase Z) (EC 3.1.26.11) (tRNA 3 endonuclease) (tRNase Z) |
| MSMEG_4814 Uncharacterized protein | MSMEG_5615 Uncharacterized protein |
| MSMEG_4817 6-phosphogluconate dehydrogenase NAD-binding protein (EC 1.1.1.-) (6-phosphogluconate dehydrogenase, NAD-binding) | MSMEG_5287 Alcohol dehydrogenase (EC 1.1.1.1) (Dehydrogenase) |
| MSMEG_4816 6-phosphogluconate dehydrogenase NAD-binding protein (EC 1.1.1.31) (6-phosphogluconate dehydrogenase, NAD-binding) | MSMEG_5286 Dihydrodipicolinate reductase (Dihydrodipicolinate reductase, N-terminus domain protein) (EC 1.17.1.8) |
| MSMEG_4811 Uncharacterized protein | MSMEG_5285 Patatin (Phospholipase, patatin family protein) |
| MSMEG_4813 Conserved domain protein (Ferredoxin) | MSMEG_2420 Uncharacterized protein |
| MSMEG_4812 Respiratory-chain NADH dehydrogenase domain, 51 kDa subunit (EC 1.6.99.5) | MSMEG_2421 OsmC family protein (Uncharacterized protein) |
| MSMEG_5506 Uncharacterized protein | MSMEG_2769 TrkA-N (TrkB protein) |
| MSMEG_6070 50S ribosomal protein L31 | MSMEG_2465 Adenine deaminase (Amidohydrolase) |
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
X4XR.rep1
dcpA.knockout.rep2
Wild.type.rep2.x
Wild.type.rep3
Biological.replicate.2.of.7..falcarinol.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Biological.replicate.6.of.6..negative.control
Biological.replicate.2.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.4.of.6..isoniazid.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X3dbfcy5plogcy3slide601norm
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
X4dbfcy5plogcy3slide901norm
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
WT_log.phase_rep2
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
dosR.WT_Replicate3
Bedaquiline.vs..DMSO.60min
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.2.of.6..negative.control
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments