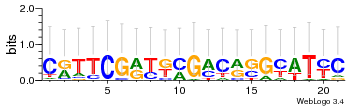
Module 386 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 2.8e-19 | 0.000000000000002 | 79 | 27 | 20160204 |
| MSMEG_6881 Transcriptional regulator, GntR family protein | MSMEG_3296 ECF-family protein sigma factor H |
| MSMEG_2570 Xanthine/uracil permease | MSMEG_3295 Uncharacterized protein |
| MSMEG_4075 CoA-binding protein | MSMEG_0916 TetR family transcriptional regulator (Transcriptional regulator, TetR family protein) |
| MSMEG_4074 Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) | MSMEG_0917 Uncharacterized protein |
| MSMEG_4593 Acyl-CoA dehydrogenase (Acyl-CoA dehydrogenase, C-terminal) (EC 1.3.8.4) | MSMEG_2569 Oxidoreductase, 2OG-Fe(II) oxygenase family protein |
| MSMEG_2100 Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase-like protein (Peptidase family protein M20/M25/M40) (EC 3.4.-.-) | MSMEG_3556 Integral membrane transporter (Putative cytosine/uracil/thiamine/allantoin permease) |
| MSMEG_2164 GntR family regulatory protein (Transcriptional regulator, putative) | MSMEG_6300 Transcriptional regulator, GntR family protein |
| MSMEG_3960 Transcriptional regulator | MSMEG_2596 Peptidase C26 (EC 4.1.3.27) |
| MSMEG_2598 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) | MSMEG_4590 Nitrilotriacetate monooxygenase component A (EC 1.14.13.-) |
| MSMEG_2599 GntR family transcriptional regulator (GntR-family protein transcriptional regulator) | MSMEG_4592 Acyl-CoA dehydrogenase (EC 1.3.99.-) (Acyl-CoA dehydrogenase family protein) |
| MSMEG_0239 O-acetylhomoserine/O-acetylserine sulfhydrylase (EC 2.5.1.49) | MSMEG_2597 Aldehyde dehydrogenase (EC 1.2.1.3) (Aldehyde dehydrogenase AldC) (EC 1.2.1.-) |
| MSMEG_0238 CoA binding domain protein (O-acetylhomoserine/O-acetylserine sulfhydrylase) | MSMEG_3959 GntR family transcriptional regulator (Transcriptional regulator) |
| MSMEG_2595 Gamma-glutamylisopropylamide synthetase (L-glutamine synthetase) (EC 6.3.1.2) | MSMEG_3958 Glyoxalase/bleomycin resistance protein/dioxygenase |
| MSMEG_3293 Uncharacterized protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.2.of.7..falcarinol.treatment
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.4.of.6..negative.control
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.2.of.6..isoniazid.treatment
Biological.replicate.5.of.6..negative.control
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
WT.with.vs..without.N_dye.swap_rep1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.2.of.6..kanamycin.treatment
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep2.x
X2XR.rep1
X2XR.rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.4.of.6..kanamycin.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments