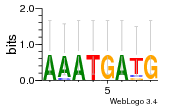
Module 420 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 0.00000021 | 0.13 | 73 | 35 | 20160204 |
| MSMEG_0909 Acyl-ACP thioesterase superfamily protein | MSMEG_5138 Nitrate reductase delta chain NarJ3 (EC 1.7.99.4) (Nitrate reductase molybdenum cofactor assembly chaperone) |
| MSMEG_1938 Uncharacterized protein | MSMEG_6903 Transcriptional regulator, PadR family protein |
| MSMEG_3684 Nuclear transport factor 2 (NTF2) domain family protein | MSMEG_4920 Probable acetyl-CoA acetyltransferase (EC 2.3.1.9) (Acetoacetyl-CoA thiolase) |
| MSMEG_1592 Uncharacterized protein | MSMEG_2264 Hydrogenase maturation protease (Peptidase M52, hydrogen uptake protein) |
| MSMEG_4015 Uncharacterized protein | MSMEG_3685 Uncharacterized protein |
| MSMEG_6548 Rieske (2Fe-2S) region (EC 1.10.9.1) (Rieske iron-sulfur protein) | MSMEG_2266 Uncharacterized protein |
| MSMEG_5441 Methionine--tRNA ligase (EC 6.1.1.10) | MSMEG_2262 Hydrogenase-2, small subunit (EC 1.12.99.6) (NADH ubiquinone oxidoreductase 20 kDa subunit) (EC 1.12.99.6) |
| MSMEG_5440 Deoxyribonuclease (TatD-related deoxyribonuclease) | MSMEG_2263 Hydrogenase-2, large subunit (EC 1.12.99.6) (Nickel-dependent hydrogenase large subunit) (EC 1.12.5.1) |
| MSMEG_5442 Glutamate dehydrogenase | MSMEG_0308 Bifunctional deaminase-reductase-like protein (Dihydrofolate reductase) |
| MSMEG_1802 ChaB protein | MSMEG_2171 L-carnitine dehydratase/bile acid-inducible protein F |
| MSMEG_0532 Transcriptional regulator, TetR family protein | MSMEG_5140 Nitrate reductase, alpha subunit (EC 1.7.99.4) |
| MSMEG_2259 Carbon starvation protein A (Carbon starvation protein CstA) | MSMEG_5141 Nitrate/nitrite transporter protein |
| MSMEG_2258 Uncharacterized protein | MSMEG_2767 Uncharacterized protein |
| MSMEG_3801 Uncharacterized protein | MSMEG_4338 Possible transcriptional regulatory protein |
| MSMEG_3681 Uncharacterized protein | MSMEG_3833 30S ribosomal protein S1 |
| MSMEG_6082 Carbonic anhydrase (EC 4.2.1.1) (Carbonate dehydratase) | MSMEG_2768 Nucleic acid binding OB-fold tRNA/helicase-type (OB-fold nucleic acid binding domain protein) |
| MSMEG_5137 Respiratory nitrate reductase, gamma subunit (EC 1.7.99.4) | MSMEG_4310 Cobalamin biosynthesis protein CobD (EC 6.3.1.10) |
| MSMEG_5139 Nitrate reductase, beta subunit (EC 1.7.99.4) (Respiratory nitrate reductase (Beta chain) narH) |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.5.of.7..panaxydol.treatment
Mycobacterium.smegmatis.Spheroplast.induced
X4XR.rep1
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..isoniazid.treatment
Bedaquiline.vs..DMSO.60min
rel.knockout.rep1
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X3dbfcy5plogcy3slide601norm
Sub.MIC.INH
Biological.replicate.1.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide901norm
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate3
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.with.vs..without.N_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Biological.replicate.4.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments