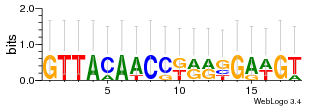
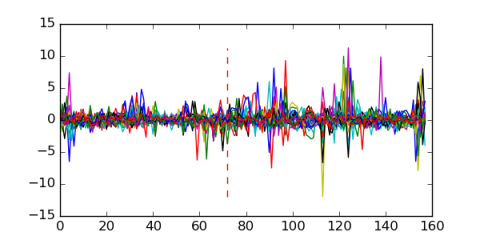
Module 445 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 670 | 1700 | 72 | 31 | 20160204 |
| MSMEG_5324 Ornithine-acylacyl carrier protein N-acyltransferase (Uncharacterized protein) | MSMEG_3253 Uncharacterized protein |
| MSMEG_5556 MFS permease | MSMEG_1964 FAD-dependent oxygenase (Mitomycin radical oxidase) (EC 1.5.3.-) |
| MSMEG_4548 Acetyltransferase, GNAT family (Acetyltransferase, gnat family protein) | MSMEG_5708 Uncharacterized protein |
| MSMEG_3014 Uncharacterized protein | MSMEG_3994 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_4640 Secreted protein | MSMEG_1655 Alpha/beta hydrolase fold protein (Hydrolase, alpha/beta fold family protein, putative) |
| MSMEG_0354 Uncharacterized protein | MSMEG_1753 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase 2 (EC 3.2.2.9) (5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase) (5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase 1) |
| MSMEG_6031 Putative short-chain type dehydrogenase/reductase MSMEG_6031/MSMEI_5872 (EC 1.1.1.-) | MSMEG_1755 Anti-sigm factor, ChrR |
| MSMEG_0604 D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding protein (EC 1.1.1.95) (Glyoxylate reductase) (EC 1.1.1.26) | MSMEG_6049 Secreted protein |
| MSMEG_0070 Uncharacterized protein | MSMEG_3089 Deoxyribose-phosphate aldolase (DERA) (EC 4.1.2.4) (2-deoxy-D-ribose 5-phosphate aldolase) (Phosphodeoxyriboaldolase) |
| MSMEG_0231 Uncharacterized protein | MSMEG_5868 Uncharacterized protein |
| MSMEG_3716 Uncharacterized protein | MSMEG_1099 Uncharacterized protein |
| MSMEG_4574 DNA-binding protein | MSMEG_2444 Dienelactone hydrolase (EC 3.1.1.45) (Dienelactone hydrolase family protein) |
| MSMEG_4633 Peptidase S9, prolyl oligopeptidase (Peptidase S9, prolyl oligopeptidase active site region) | MSMEG_4339 Tetratricopeptide repeat family protein (Transcriptional regulator, LuxR family) |
| MSMEG_2529 D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding protein (EC 1.1.1.26) (Glyoxylate reductase) | MSMEG_4848 Uncharacterized protein |
| MSMEG_2528 Glycerate kinase (EC 2.7.1.31) | MSMEG_2911 Integral membrane transport protein (Major facilitator superfamily MFS_1) |
| MSMEG_1733 Uncharacterized protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.30min
Wild.type.rep3
Biological.replicate.4.of.6..kanamycin.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.7.of.7..falcarinol.treatment
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
MB100pruR.vs..MB100.rep.2
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
Sub.MIC.INH
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
X2.5..vs..50..air.saturation.Replicate.4
Biological.replicate.4.of.7..ethambutol.treatment
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
Biological.replicate.5.of.6..negative.control
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
X2XR.rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments