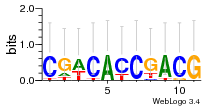
Module 450 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.000000000025 | 0.0059 | 84 | 26 | 20160204 |
| MSMEG_3640 Malate synthase G (EC 2.3.3.9) | MSMEG_6877 Branched-chain amino acid transport system ATP-binding protein |
| MSMEG_6880 Hydrophobic amino acid ABC transporter, putative | MSMEG_6876 Branched chain amino acid transport ATP-binding protein (Branched-chain amino acid ABC transporter ATP-binding protein) (EC 3.6.3.27) |
| MSMEG_0478 Secreted protein, putative | MSMEG_6879 ABC-type branched-chain amino acid transport system, permease protein I (Integral membrane protein of the ABC-type Nat permease for neutral amino acids NatD) |
| MSMEG_3117 Cytochrome aa3 controlling protein (Unidentified antibiotic-transport integral membrane ABC transporter) | MSMEG_6878 Branched-chain amino acid ABC transporter permease protein (Inner-membrane translocator) |
| MSMEG_2089 Cell division ATP-binding protein FtsE | MSMEG_2091 SsrA-binding protein (Small protein B) |
| MSMEG_0475 Nucleotide-sugar dehydrogenase | MSMEG_2090 Cell division protein FtsX |
| MSMEG_4687 Cytosine deaminase (EC 3.5.4.1) | MSMEG_6906 Alpha/beta hydrolase fold protein (Putative hydrolase) |
| MSMEG_1570 Carboxylesterase | MSMEG_3638 CBS domain protein (Conserved hypothetical membrane protein) |
| MSMEG_4685 Oxidoreductase | MSMEG_0476 Chitin synthase |
| MSMEG_0050 Uncharacterized protein | MSMEG_6202 Secreted protein |
| MSMEG_3637 CBS domain protein | MSMEG_6203 Cysteine synthase/cystathionine beta-synthase (EC 2.5.1.47) (Cysteine synthase/cystathionine beta-synthase family protein) |
| MSMEG_4248 1-acylglycerol-3-phosphate O-acyltransferase (EC 2.3.1.51) | MSMEG_5284 Patatin |
| MSMEG_3639 Uncharacterized protein | MSMEG_0049 Pirin domain protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
dcpA.knockout.rep2
M..smegmatis.Acid.NO.treated
X4XR.rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.2.of.7..falcarinol.treatment
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
X2XR.rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Wild.type.rep1.y
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DamtR.N.surplus_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
Sub.MIC.INH
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
WT_log.phase_rep2
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
stphcy5plogcy3slide487norm
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.2.of.7..ethambutol.treatment
dosR.WT_Replicate2
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.60min
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.7.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DamtR.N.surplus_dyeswap_rep2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
X4dbfcy3plogcy5slide903norm
Wild.type.rep3
Comments
Log in to post comments