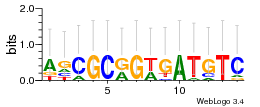
Module 457 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 600 | 1700 | 72 | 27 | 20160204 |
| MSMEG_3642 Glycine dehydrogenase (decarboxylating) (EC 1.4.4.2) (Glycine cleavage system P-protein) (Glycine decarboxylase) (Glycine dehydrogenase (aminomethyl-transferring)) | MSMEG_3651 Membrane associated protein (Uncharacterized protein) |
| MSMEG_6093 Uncharacterized protein | MSMEG_5336 Amidate substrates transporter protein (Putative transporter protein amiS2) |
| MSMEG_6091 ATP-dependent Clp protease ATP-binding subunit ClpC1 | MSMEG_5562 Ribonuclease H (RNase H) (EC 3.1.26.4) |
| MSMEG_5488 Response regulator MprA (Mycobacterial persistence regulator A) | MSMEG_1611 Putative transcriptional regulatory protein (Transcriptional regulator, TetR family protein, putative) |
| MSMEG_6734 Dibenzothiophene desulfurization enzyme A (Putative desulfurization/monooxygenase) (EC 1.14.14.5) | MSMEG_3432 Choline dehydrogenase (EC 1.1.99.1) (Glucose-methanol-choline oxidoreductase) |
| MSMEG_1060 Putative Lsr2 protein (Uncharacterized protein) | MSMEG_0657 Rieske 2Fe-2S domain protein |
| MSMEG_5778 Alcohol dehydrogenase, zinc-binding (Alcohol dehydrogenase, zinc-binding protein) (EC 1.6.5.5) | MSMEG_5435 Putative ligase MSMEG_5435/MSMEI_5285 (EC 6.2.1.-) |
| MSMEG_0296 Transcriptional regulator, MarR family (Transcriptional regulator, MarR family protein) | MSMEG_2321 |
| MSMEG_0290 Diacylglycerol O-acyltransferase (EC 2.3.1.20) | MSMEG_1610 GMP synthase [glutamine-hydrolyzing] (EC 6.3.5.2) (GMP synthetase) (Glutamine amidotransferase) |
| MSMEG_1997 D-amino acid aminohydrolase (EC 3.5.1.82) (N-acyl-D-glutamate amidohydrolase) | MSMEG_5487 Signal transduction histidine-protein kinase/phosphatase MprB (EC 2.7.13.3) (EC 3.1.3.-) (Mycobacterial persistence regulator B) |
| MSMEG_2954 LacI-family protein transcriptional regulator (Transcriptional regulator, LacI family) (EC 5.1.1.1) | MSMEG_6578 Uncharacterized protein |
| MSMEG_5852 Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase) | MSMEG_3939 Universal stress protein family protein (UspA) |
| MSMEG_1268 NUDIX hydrolase (Nudix hydrolase) | MSMEG_0385 Putative glycosyl transferase (EC 2.-.-.-) |
| MSMEG_3715 Linear gramicidin synthetase subunit C (EC 5.1.1.-) (Non-ribosomal peptide synthase) (EC 1.2.1.31) |
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate3
dosR.WT_Replicate1
Biological.replicate.2.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..isoniazid.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Plogcy5plogcy3slide113norm
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
X4dbfcy5plogcy3slide480norm
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
WT_log.phase_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.3.of.6..kanamycin.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.with.vs..without.N_rep2
WT.vs..DamtR.N.surplus_.dyeswap_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
WT.vs..DamtR.N.surplus_rep2
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments