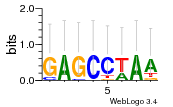
Module 462 Residual: 0.56
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.56 | 0.0000000000022 | 210 | 79 | 34 | 20160204 |
| MSMEG_6171 Uncharacterized protein | MSMEG_6899 Conserved transmembrane protein (Uncharacterized protein) |
| MSMEG_6932 Uncharacterized protein | MSMEG_6290 DNA-binding protein (Putative DNA-binding protein) |
| MSMEG_4014 Amidase, hydantoinase/carbamoylase (EC 3.5.1.87) (N-carbamoyl-L-amino acid amidohydrolase) | MSMEG_6900 Penicillin-binding protein 1A |
| MSMEG_4016 Uncharacterized protein | MSMEG_6169 Type II secretion system protein E (Type II/IV secretion system protein) |
| MSMEG_1049 Methyltransferase type 11 (Methyltransferase, UbiE/COQ5 family protein) | MSMEG_4006 Transcriptional regulator, CdaR, putative |
| MSMEG_0681 P450 heme-thiolate protein | MSMEG_5700 Secreted protein (Transglycosylase domain protein) |
| MSMEG_0682 Uncharacterized protein | MSMEG_2962 Protein-export membrane protein SecF |
| MSMEG_0683 Uncharacterized protein | MSMEG_4829 Cytochrome P450 |
| MSMEG_5638 Metallo-beta-lactamase family protein (Zn-dependent hydrolase of the beta-lactamase fold-like protein) | MSMEG_3596 AAA ATPase (ATPase) |
| MSMEG_5639 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) | MSMEG_6242 Alcohol dehydrogenase, iron-containing (EC 1.1.1.1) (Iron-containing alcohol dehydrogenase) (EC 1.1.1.202) |
| MSMEG_0076 Antigen MTB48 (Uncharacterized protein) | MSMEG_6240 Uncharacterized protein |
| MSMEG_3595 Uncharacterized protein | MSMEG_6241 ATPase associated with various cellular activities AAA_5 (ATPase associated with various cellular activities, AAA-5) |
| MSMEG_3499 Putative exported protein (Uncharacterized protein) | MSMEG_2035 Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Putative flavin-containing monoamine oxidase aofH) |
| MSMEG_2961 Protein translocase subunit SecD | MSMEG_1695 Phosphoglucomutase/phosphomannomutase (Phosphoglucomutase/phosphomannomutase alpha/beta/alpha domain I) (EC 5.4.2.8) |
| MSMEG_3615 Zinc-binding alcohol dehydrogenase family protein (EC 1.-.-.-) | MSMEG_6222 Integral membrane protein (RDD domain containing protein) |
| MSMEG_2731 Uncharacterized protein MSMEG_2731/MSMEI_2664 | MSMEG_1136 Uncharacterized protein |
| MSMEG_2964 Adenine phosphoribosyltransferase (APRT) (EC 2.4.2.7) | MSMEG_3766 Uncharacterized protein |
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.4.of.6..negative.control
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.4.of.6..isoniazid.treatment
dosR.WT_Replicate3
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
WT.vs..DamtR.N.surplus_dyeswap_rep2
DglnR_without.vs..with.N_rep1
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
DamtR.with.vs..DamtR.without.N_.rep2
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
X4dbfcy5plogcy3slide480norm
X3dbfcy5plogcy3slide603norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
WT.vs..DamtR.N.surplus_rep2
Comments
Log in to post comments