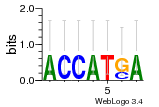
Module 464 Residual: 0.41
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.41 | 26 | 780 | 83 | 20 | 20160204 |
| MSMEG_0749 Carbon monoxide dehydrogenase subunit G (Carbon monoxide dehydrogenase subunit G (CoxG) family protein) | MSMEG_0747 Carbon monoxide dehydrogenase F protein |
| MSMEG_0748 ATPase associated with various cellular activities, AAA_5 | MSMEG_1148 MCE-family protein Mce6F (Mce related protein) |
| MSMEG_1679 AmiB (Amidohydrolase AmiB1) | MSMEG_0744 Carbon monoxide dehydrogenase medium chain (Carbon monoxide dehydrogenase, medium subunit) (EC 1.2.99.2) |
| MSMEG_1677 Aspartate ammonia-lyase (EC 4.3.1.1) | MSMEG_1144 MCE-family protein Mce6B (Virulence factor Mce family protein) |
| MSMEG_0751 Uncharacterized protein (VWA containing CoxE-like protein) | MSMEG_1145 MCE-family protein Mce6C (Virulence factor Mce family protein) |
| MSMEG_0745 Carbon monoxide dehydrogenase (Small chain), CoxS (EC 1.2.99.2) ([2Fe-2S] binding domain protein) | MSMEG_1146 Virulence factor Mce family protein |
| MSMEG_1682 Flavin-containing monooxygenase FMO | MSMEG_1147 Mce family protein, Mce5E (Mce related protein) |
| MSMEG_1681 Endoribonuclease L-PSP superfamily protein | MSMEG_1141 ABC-transporter integral membrane protein (YrbE family protein, YrbE5A) |
| MSMEG_1680 Uncharacterized protein | MSMEG_1142 ABC-transporter integral membrane protein (Conserved hypothetical integral membrane protein YrbE6B) |
| MSMEG_0746 Carbon monoxide dehydrogenase (Large chain), CoxL (EC 1.2.99.2) (Carbon-monoxide dehydrogenase, large subunit) | MSMEG_1143 MCE-family protein Mce6A (Mce related protein) |
mc2155.vs.Dcrp1.replicate.1
Bedaquiline.vs..DMSO.30min
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.2.of.7..panaxydol.treatment
dcpA.knockout.rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate2
KO_log.phase_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.6..negative.control
Biological.replicate.6.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep2
WT.vs..DglnR.N_starvation_dye.swap_rep2
Biological.replicate.3.of.7..ethambutol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Biological.replicate.5.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
Sub.MIC.INH
Plogcy5plogcy3slide111norm
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.5
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X4XR.rep1
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Biological.replicate.1.of.7..falcarinol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
Biological.replicate.5.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
WT.vs..DglnR.N_starvation_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments