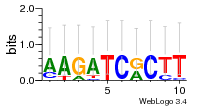
Module 466 Residual: 0.53
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.53 | 0.00002 | 0.000044 | 78 | 29 | 20160204 |
| MSMEG_5045 D-2-hydroxyglutarate dehydrogenase (EC 1.1.99.-) (Putative glyoclate oxidase FAD-linked subunit) | MSMEG_4109 NAD(P) transhydrogenase, alpha subunit (EC 1.6.1.1) |
| MSMEG_5044 ATPase (Uncharacterized protein) | MSMEG_4107 Phosphoglycerate mutase (EC 5.4.2.-) (Phosphoglycerate mutase, putative) |
| MSMEG_3074 Uncharacterized protein | MSMEG_1778 Uncharacterized protein |
| MSMEG_3072 Riboflavin biosynthesis protein RibBA | MSMEG_5310 SAM-dependent methyltransferase |
| MSMEG_3073 6,7-dimethyl-8-ribityllumazine synthase (DMRL synthase) (LS) (Lumazine synthase) (EC 2.5.1.78) | MSMEG_1589 Uncharacterized protein |
| MSMEG_1490 3-oxoacyl-ACP synthase III | MSMEG_1420 Probable transcriptional regulatory protein (Transcriptional regulator, TetR family) |
| MSMEG_1648 Putative transcriptional regulator | MSMEG_1955 Uncharacterized protein |
| MSMEG_5400 Dehydrogenase (FAD dependent oxidoreductase) | MSMEG_6221 Integral membrane protein (Putative conserved transmembrane protein) |
| MSMEG_5816 Uncharacterized protein | MSMEG_1587 Alpha/beta hydrolase fold (Alpha/beta hydrolase fold protein) |
| MSMEG_1453 Citrate transporter (Putative integral membrane protein) | MSMEG_2919 2-dehydropantoate 2-reductase (Ketopantoate reductase ApbA/PanE-like protein) (EC 1.1.1.169) |
| MSMEG_6858 Epoxide hydrolase (Epoxide hydrolase 1) (EC 3.3.2.9) | MSMEG_4275 Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase (NN:DBI PRT) (EC 2.4.2.21) (N(1)-alpha-phosphoribosyltransferase) |
| MSMEG_6850 Alpha/beta hydrolase (Alpha/beta hydrolase fold protein) (EC 3.3.2.9) | MSMEG_4274 Cobinamide kinase / cobinamide phosphate guanyltransferase (EC 2.7.1.156) (EC 2.7.7.62) |
| MSMEG_5076 ABC transporter membrane-spanning protein (Putative ABC transporter membrane-spanning protein) | MSMEG_4277 Adenosylcobinamide-GDP ribazoletransferase (EC 2.7.8.26) (Cobalamin synthase) (Cobalamin-5'-phosphate synthase) |
| MSMEG_5077 Conserved integral membrane protein (Probable conserved integral membrane protein) | MSMEG_3261 Uncharacterized protein |
| MSMEG_1489 Uncharacterized protein |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
dcpA.knockout.rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.30min
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Biological.replicate.1.of.7..panaxydol.treatment
dosR.WT_Replicate1
KO_log.phase_rep2
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Wild.type.rep1.x
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
stphcy3plogcy5slide581norm
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.3.of.6..kanamycin.treatment
Biological.replicate.4.of.7..ethambutol.treatment
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Wild.type.rep2.x
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Wild.type.rep3
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
X2XR.rep1
X2XR.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments