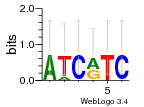
Module 470 Residual: 0.47
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.47 | 4.9e-23 | 0.2 | 80 | 29 | 20160204 |
| MSMEG_4792 Virulence factor Mce family protein (Virulence factor mce family protein) | MSMEG_3802 Tetratricopeptide TPR_4 (Tetratricopeptide repeat family protein) |
| MSMEG_4793 Virulence factor Mce family protein | MSMEG_2968 Uncharacterized protein |
| MSMEG_4794 Virulence factor (Virulence factor Mce family protein) | MSMEG_2967 Conserved protein/domain typically associated with flavoprotein oxygenase DIM6/NTAB family-like protein (Uncharacterized protein) |
| MSMEG_0960 Putative transmembrane protein (Uncharacterized protein) | MSMEG_2966 Major facilitator family protein transporter |
| MSMEG_0648 Uncharacterized protein | MSMEG_0957 Uncharacterized protein |
| MSMEG_6567 Iron-dependent peroxidase | MSMEG_0958 Uncharacterized protein |
| MSMEG_3789 Inner membrane permease YgbN (Low-affinity gluconate transporter) | MSMEG_0959 Putative conserved transmembrane protein |
| MSMEG_4131 Uncharacterized protein | MSMEG_0628 Amidohydrolase 2 (Amidohydrolase family protein) |
| MSMEG_1022 Uncharacterized protein | MSMEG_1945 Ion channel membrane protein (Transmembrane cation transporter) |
| MSMEG_6569 Putative secreted protein (Uncharacterized protein) | MSMEG_1946 NADH pyrophosphatase (EC 3.6.1.22) (NADH pyrophosphatase NudC) |
| MSMEG_3787 D-Aminoacylase (EC 3.5.1.-) (D-aminoacylase) (EC 3.5.1.81) | MSMEG_2302 |
| MSMEG_0093 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_0093 (EC 2.1.1.-) | MSMEG_0095 Putative S-adenosyl-L-methionine-dependent methyltransferase MSMEG_0095/MSMEI_0092 (EC 2.1.1.-) |
| MSMEG_0629 Trans-aconitate 2-methyltransferase (EC 2.1.1.144) | MSMEG_3788 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase (EC 4.1.3.16) (KHG/KDPG aldolase) (EC 4.1.2.14) |
| MSMEG_5698 Cyclic pyranopterin monophosphate synthase (EC 4.1.99.18) (Molybdenum cofactor biosynthesis protein A) | MSMEG_3107 Uncharacterized protein |
| MSMEG_5699 ThiS family protein |
KO_stationary.phase_rep1
WT.vs..DamtR.without.N_rep1
KO_log.phase_rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
Plogcy5plogcy3slide110norm
Biological.replicate.3.of.7..ethambutol.treatment
dosR.WT_Replicate3
KO_log.phase_rep2
Wild.type.rep2.y
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep2
rel.knockout.rep1
Biological.replicate.1.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
X0.6..vs..50..air.saturation.Replicate.1
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.6.of.7..ethambutol.treatment
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Plogcy5plogcy3slide113norm
Biological.replicate.7.of.7..panaxydol.treatment
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide603norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.5
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
Biological.replicate.4.of.6..isoniazid.treatment
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep2.x
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments