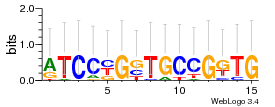
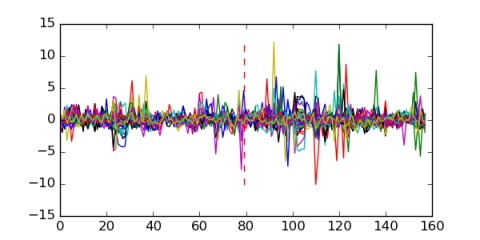
Module 491 Residual: 0.54
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.54 | 0.0000000000000058 | 0.0000018 | 79 | 34 | 20160204 |
| MSMEG_5485 Molybdopterin biosynthesis protein | MSMEG_0059 ATPase, AAA family protein |
| MSMEG_5486 Peptidase S1 and S6, chymotrypsin/Hap (Serine protease PepD) | MSMEG_0058 Uncharacterized protein |
| MSMEG_0868 Uncharacterized protein | MSMEG_3653 Uncharacterized protein |
| MSMEG_0869 Uncharacterized protein | MSMEG_6439 Putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase (HMG aldolase) (EC 4.1.3.17) (Oxaloacetate decarboxylase) (OAA decarboxylase) (EC 4.1.1.3) (Regulator of ribonuclease activity homolog) (RraA-like protein) |
| MSMEG_6394 Cutinase (Uncharacterized protein) | MSMEG_0810 Putative pyrimidine permease RutG (Uracil-xanthine permease) |
| MSMEG_6395 Uncharacterized protein | MSMEG_3799 Uncharacterized protein |
| MSMEG_4754 K+-dependent Na+/Ca+ exchanger related-protein (Na+/Ca+ antiporter, CaCA family) | MSMEG_2084 Uncharacterized protein |
| MSMEG_0804 Uncharacterized protein | MSMEG_4424 Endoribonuclease L-PSP |
| MSMEG_2083 Inositol monophosphatase (Putative histidinol-phosphate phosphatase, inositol monophosphatase family) (EC 3.1.3.15) | MSMEG_2110 Uncharacterized protein |
| MSMEG_5594 Ferredoxin-dependent glutamate synthase (EC 1.4.1.13) | MSMEG_6202 Secreted protein |
| MSMEG_5593 Pyruvate dehydrogenase (Thiamine pyrophosphate enzyme-like TPP binding protein) (EC 1.2.3.3) | MSMEG_6203 Cysteine synthase/cystathionine beta-synthase (EC 2.5.1.47) (Cysteine synthase/cystathionine beta-synthase family protein) |
| MSMEG_5799 Nucleoside-diphosphate-sugar epimerase | MSMEG_0062 Cell division FtsK/SpoIIIE (Ftsk/spoiiie family protein) |
| MSMEG_5798 Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIR synthase) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) | MSMEG_0060 Putative conserved membrane protein (Uncharacterized protein) |
| MSMEG_3800 Uncharacterized protein | MSMEG_2700 Uncharacterized protein |
| MSMEG_1769 Uncharacterized protein (UsfY protein) | MSMEG_1131 Tryptophan-rich sensory protein |
| MSMEG_0057 Uncharacterized protein | MSMEG_0061 Ftsk/spoiiie family protein (Uncharacterized protein) |
| MSMEG_0056 Uncharacterized protein | MSMEG_1215 Serine/threonine-protein kinase PknE, putative (EC 2.7.11.1) |
Biological.replicate.2.of.7..panaxydol.treatment
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
rel.knockout.rep2
Wild.type.rep3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
KO_log.phase_rep2
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
DglnR_without.vs..with.N_rep1
MB100pruR.vs..MB100.rep.4
DamtR.with.vs..DamtR.without.N_.rep2
MB100pruR.vs..MB100.rep.2
DglnR_without.vs..with.N_rep2
mc2155.vs.Dcrp1.replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
stphcy5plogcy3slide487norm
Wild.type.rep2.y
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.2.of.6..negative.control
WT_stationary.phase_rep1
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Bedaquiline.vs..DMSO.60min
Biological.replicate.2.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
X4dbfcy5plogcy3slide480norm
Biological.replicate.5.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
Wild.type.rep1.y
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Biological.replicate.4.of.6..kanamycin.treatment
BatchCulture_LBTbroth_StationaryPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
dosR.WT_Replicate2
Comments
Log in to post comments