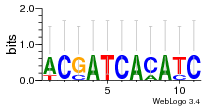
Module 502 Residual: 0.50
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.50 | 0.00000000023 | 120 | 85 | 17 | 20160204 |
| MSMEG_2703 Hydrogenase assembly chaperone HypC/HupF (Hydrogenase maturation factor, HypC) | MSMEG_1715 L-arabinose isomerase (EC 5.3.1.4) |
| MSMEG_5999 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_1714 L-ribulose-5-phosphate 4-epimerase (EC 5.1.3.4) (L-ribulose-5-phosphate 4-epimerase UlaF) |
| MSMEG_2702 Hydrogenase expression/formation protein HypD (Hydrogenase expression/formation protein hypD) | MSMEG_6002 Coenzyme A transferase, subunit A (Putative coa-transferase (Alpha subunit)) (EC 2.8.3.12) |
| MSMEG_6000 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_6003 Coenzyme A transferase, subunit B |
| MSMEG_6008 Acetyl-CoA acetyltransferase (Putative acetyl-COA acetyltransferase Fada6 (Acetoacetyl-COA thiolase)) (EC 2.3.1.-) | MSMEG_6004 2-nitropropane dioxygenase NPD (EC 1.13.12.16) (Oxidoreductase, 2-nitropropane dioxygenase family protein) |
| MSMEG_6009 HTH-type transcriptional repressor KstR2 | MSMEG_6012 Putative acyl-CoA dehydrogenase |
| MSMEG_2854 Putative integral membrane protein yrbE4B (Uncharacterized protein) | MSMEG_6011 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) |
| MSMEG_1713 Ribulokinase (EC 2.7.1.16) | MSMEG_2858 Virulence factor mce family protein |
| MSMEG_6001 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) |
KO_stationary.phase_rep1
WT.vs..DglnR.N_starvation_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.4.of.6..negative.control
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.3.of.7..ethambutol.treatment
dosR.WT_Replicate3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.3.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Biological.replicate.1.of.7..panaxydol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
stphcy5plogcy3slide487norm
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.2.of.6..negative.control
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
mc2155.vs.Dcrp1.replicate.3
Slow.vs..Fast.Replicate.4
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.60min
Biological.replicate.2.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Biological.replicate.3.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.with.vs..without.N_rep2
Wild.type.rep1.x
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate1
dosR.WT_Replicate2
Plogcy5plogcy3slide113norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments