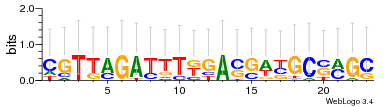
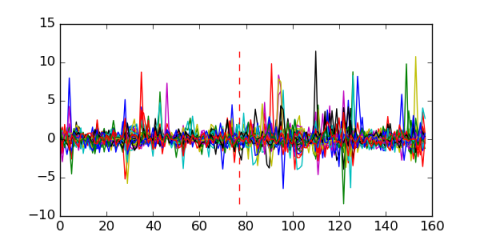
Module 507 Residual: 0.51
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.51 | 5e-30 | 0.000064 | 77 | 31 | 20160204 |
| MSMEG_2950 Carbon monoxide dehydrogenase medium chain (EC 1.2.99.2) (Carbon monoxide dehydrogenase, medium chain) | MSMEG_5073 Putative O-methyltransferase MSMEG_5073/MSMEI_4947 (EC 2.1.1.-) |
| MSMEG_3963 Uncharacterized protein | MSMEG_2949 Aldehyde oxidase and xanthine dehydrogenase molybdopterin binding protein (EC 1.17.1.4) (Carbon-monoxide dehydrogenase) |
| MSMEG_5487 Signal transduction histidine-protein kinase/phosphatase MprB (EC 2.7.13.3) (EC 3.1.3.-) (Mycobacterial persistence regulator B) | MSMEG_5524 Succinyl-CoA ligase [ADP-forming] subunit alpha (EC 6.2.1.5) |
| MSMEG_5488 Response regulator MprA (Mycobacterial persistence regulator A) | MSMEG_5525 Succinyl-CoA ligase [ADP-forming] subunit beta (EC 6.2.1.5) (Succinyl-CoA synthetase subunit beta) (SCS-beta) |
| MSMEG_3105 Protoheme IX farnesyltransferase (EC 2.5.1.-) (Heme B farnesyltransferase) (Heme O synthase) | MSMEG_6642 Caib/baif family protein (L-carnitine dehydratase/bile acid-inducible protein F) (EC 2.8.3.16) |
| MSMEG_6639 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein, putative) | MSMEG_5561 HPP family protein |
| MSMEG_5348 Medium chain fatty-acid-CoA ligase FadD14 (EC 2.3.1.86) (Medium-chain fatty acid-CoA ligase) | MSMEG_5451 Alkylated DNA repair protein |
| MSMEG_2951 (2Fe-2S)-binding protein (EC 1.2.99.2) ([2Fe-2S] binding domain protein) | MSMEG_4129 TetR family transcriptional regulator (Uncharacterized protein) |
| MSMEG_4158 Putative transposase (Uncharacterized protein) | MSMEG_4128 Uncharacterized protein |
| MSMEG_1749 Luciferase-like protein (EC 1.14.14.5) (Putative monooxygenase) | MSMEG_1752 Uncharacterized protein |
| MSMEG_1106 AraC family transcriptional regulator (Transcriptional regulator, AraC family protein) | MSMEG_6641 Nitrilotriacetate monooxygenase component A (EC 1.14.13.-) (Xenobiotic compound monooxygenase, DszA family, A subunit) |
| MSMEG_1107 Isonitrile hydratase, putative (ThiJ/PfpI) | MSMEG_0482 Dihydroxy-acid dehydratase (EC 4.2.1.9) (Dihydroxyacid dehydratase) (EC 4.2.1.9) |
| MSMEG_5399 ATP-dependent DNA helicase RecQ (EC 3.6.1.-) | MSMEG_0480 Transcriptional regulator, GntR family (Transcriptional regulator, GntR family protein) |
| MSMEG_0520 Porin MspB | MSMEG_0481 FAD dependent oxidoreductase |
| MSMEG_5496 MscS Mechanosensitive ion channel | MSMEG_4276 Branched-chain-amino-acid aminotransferase (BCAT) (EC 2.6.1.42) |
| MSMEG_5072 ECF RNA polymerase sigma factor SigE (ECF sigma factor SigE) (Alternative RNA polymerase sigma factor SigE) (RNA polymerase sigma-E factor) (Sigma-E factor) |
Wild.type.rep2.x
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
WT.vs..DamtR.without.N_rep2
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.5.of.6..negative.control
DglnR_without.vs..with.N_rep1
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep1
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
Biological.replicate.5.of.6..kanamycin.treatment
Biological.replicate.1.of.7..panaxydol.treatment
X3dbfcy5plogcy3slide603norm
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
WT_log.phase_rep2
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.1
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
dosR.WT_Replicate2
WT.vs..DglnR.N_starvation_rep1
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
Wild.type.rep1.y
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments