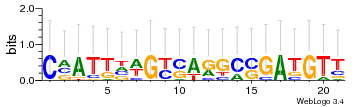
Module 521 Residual: 0.48
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.48 | 0.0000000000006 | 0.000087 | 74 | 33 | 20160204 |
| MSMEG_2414 Phosphopantetheine adenylyltransferase (EC 2.7.7.3) (Dephospho-CoA pyrophosphorylase) (Pantetheine-phosphate adenylyltransferase) (PPAT) | MSMEG_5640 Amylo-alpha-1,6-glucosidase |
| MSMEG_4034 NAD dependent epimerase/dehydratase family protein (NmrA-like family protein) | MSMEG_5499 Uncharacterized protein |
| MSMEG_4135 Possible lysine decarboxylase superfamily protein (Uncharacterized protein) | MSMEG_5523 Peptidase (Peptidase S9, prolyl oligopeptidase active site region) (EC 3.4.19.1) |
| MSMEG_1824 Transcriptional regulator, LytR family protein | MSMEG_3362 Enoyl-CoA hydratase (Enoyl-CoA hydratase/isomerase) (EC 4.2.1.17) |
| MSMEG_3566 FadD16 (FadD16 protein) | MSMEG_3363 Regulatory protein, TetR (Transcriptional regulator, TetR family) |
| MSMEG_0439 UPF0324 membrane protein (Uncharacterized protein) | MSMEG_4003 Uncharacterized protein |
| MSMEG_6357 Uncharacterized protein | MSMEG_3364 Lysine exporter protein (LYSE/YGGA) (RhtB family protein transporter) |
| MSMEG_6198 DNA-binding protein, putative (Purine catabolism PurC domain protein) | MSMEG_5947 Serine O-acetyltransferase (EC 2.3.1.30) |
| MSMEG_2792 ATP-dependent protease ATP-binding subunit ClpC2 (Clp amino terminal domain protein) | MSMEG_1567 Uncharacterized protein |
| MSMEG_2416 Uncharacterized protein | MSMEG_6060 Permease |
| MSMEG_6059 IrpA (Uncharacterized protein) | MSMEG_6061 Uncharacterized protein |
| MSMEG_6058 Cadmium transporting P-type ATPase (Metal cation transporting P-type ATPase CtpC) | MSMEG_1568 Glutamine--fructose-6-phosphate aminotransferase [isomerizing] (EC 2.6.1.16) (D-fructose-6-phosphate amidotransferase) (GFAT) (Glucosamine-6-phosphate synthase) (Hexosephosphate aminotransferase) (L-glutamine--D-fructose-6-phosphate amidotransferase) |
| MSMEG_2840 Putative conserved membrane protein (Uncharacterized protein) | MSMEG_2427 Bifunctional uridylyltransferase/uridylyl-removing enzyme (UTase/UR) (Bifunctional [protein-PII] modification enzyme) (Bifunctional nitrogen sensor protein) |
| MSMEG_3945 Universal stress protein family protein | MSMEG_4427 Drug resistance transporter, EmrB/QacA family (Transmembrane efflux pump) |
| MSMEG_5037 Oxidoreductase, FAD-binding (Putative glycolate oxidase FAD-linked subunit) (EC 1.1.2.4) | MSMEG_3915 NAD-dependent alcohol dehydrogenase (EC 1.1.1.1) |
| MSMEG_3715 Linear gramicidin synthetase subunit C (EC 5.1.1.-) (Non-ribosomal peptide synthase) (EC 1.2.1.31) | MSMEG_5376 Uncharacterized protein |
| MSMEG_5642 Acetyl-coenzyme a carboxylase carboxyl transferase (Putative carboxyltransferase subunit of acetyl-CoA carboxylase) (EC 6.4.1.2) |
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
Biological.replicate.4.of.6..negative.control
Sub.MIC.INH
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.4.of.7..ethambutol.treatment
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
KO_log.phase_rep2
KO_log.phase_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.1.of.6..kanamycin.treatment
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
Biological.replicate.3.of.6..negative.control
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.5.of.6..negative.control
X2.5..vs..50..air.saturation.Replicate.4
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
stphcy3plogcy5slide581norm
WT.with.vs..without.N_dye.swap_rep1
X2.5..vs..50..air.saturation.Replicate.2
WT.with.vs..without.N_dye.swap_rep2
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
Biological.replicate.2.of.6..kanamycin.treatment
Slow.vs..Fast.Replicate.1
WT.vs..DglnR.N_starvation_dye.swap_rep2
Plogcy5plogcy3slide110norm
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.5.of.6..isoniazid.treatment
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.5.of.6..kanamycin.treatment
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments