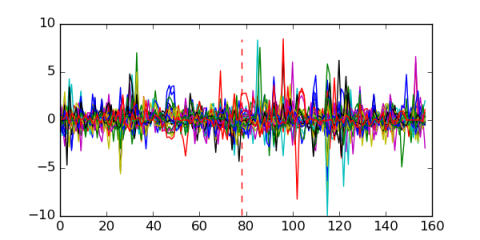
Module 523 Residual: 0.60
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.60 | 1.6e-17 | 0.000000000014 | 78 | 31 | 20160204 |
| MSMEG_6097 Pantothenate synthetase (PS) (EC 6.3.2.1) (Pantoate--beta-alanine ligase) (Pantoate-activating enzyme) | MSMEG_5470 Molybdopterin biosynthesis protein MoeA 1 (Molybdopterin biosynthesis protein MoeA1) |
| MSMEG_6934 Thioredoxin | MSMEG_2580 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (flavodoxin) (EC 1.17.7.3) (1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase) |
| MSMEG_5732 Monooxygenase | MSMEG_4693 Uncharacterized protein |
| MSMEG_6154 Adenylate cyclase, family protein 3 (Adenylate/guanylate cyclase) (EC 4.6.1.1) | MSMEG_4692 Uncharacterized protein MSMEG_4692/MSMEI_4575 |
| MSMEG_6153 DNA polymerase III subunit delta (EC 2.7.7.7) | MSMEG_1393 Conserved threonine rich protein (Uncharacterized protein) |
| MSMEG_6098 Chalcone/stilbene synthase family protein (Conserved alanine and leucine rich protein) | MSMEG_1395 Lipoprotein, putative |
| MSMEG_3784 Transcriptional regulator IclR (Transcriptional regulator, IclR family protein) | MSMEG_1394 Conserved transmembrane protein (Probable conserved transmembrane protein) |
| MSMEG_4054 Uncharacterized protein | MSMEG_1396 LprB protein |
| MSMEG_3782 Endoribonuclease L-PSP (Endoribonuclease L-PSP, putative) | MSMEG_3977 Luciferase-like protein (EC 1.5.99.11) (Uncharacterized protein) |
| MSMEG_3986 Acetyl-coenzyme A synthetase (EC 6.2.1.1) (Putative acetate--CoA ligase) | MSMEG_3976 DNA-binding protein (Transcriptional regulator, TetR family) |
| MSMEG_0457 Topoisomerase subunit TopoN (EC 5.99.1.3) (DNA gyrase subunit B-like protein MSMEG_0457/MSMEI_0444) | MSMEG_0756 Putative zinc metalloprotease Rip2 (EC 3.4.24.-) |
| MSMEG_0456 Topoisomerase subunit TopoM (EC 5.99.1.3) | MSMEG_0719 Flavohemoprotein |
| MSMEG_0459 Putative two-component system response regulator, LuxR family (Two-component system response regulator) | MSMEG_5166 Na+/solute symporter (SSS sodium solute transporter superfamily) |
| MSMEG_0458 Two-component system sensor kinase | MSMEG_5167 Major facilitator superfamily protein MFS_1 |
| MSMEG_1450 Uncharacterized protein | MSMEG_5165 Integral membrane protein |
| MSMEG_1265 Uncharacterized protein |
KO_stationary.phase_rep1
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Bedaquiline.vs..DMSO.15min
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate4
Biological.replicate.1.of.6..negative.control
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Biological.replicate.2.of.6..negative.control
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep1
Biological.replicate.7.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide601norm
Wild.type.rep1.y
MB100pruR.vs..MB100.rep.4
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
X0.6..vs..50..air.saturation.Replicate.4
Biological.replicate.2.of.7..falcarinol.treatment
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Wild.type.rep2.y
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
WT.with.vs..without.N_dye.swap_rep1
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
X4XR.rep1
Slow.vs..Fast.Replicate.4
WT.with.vs..without.N_dye.swap_rep2
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep2
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
Wild.type.rep3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.with.vs..without.N_rep1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
X2XR.rep2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.vs..DglnR.N_starvation_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments