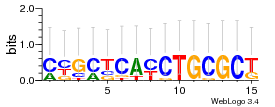
Module 533 Residual: 0.59
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.59 | 1.4e-17 | 0.000036 | 81 | 24 | 20160204 |
| MSMEG_0860 CDP-diacylglycerol--serine O-phosphatidyltransferase (EC 2.7.8.8) | MSMEG_1901 DNA-binding HTH domain containing protein, putative (Transcriptional regulator, LuxR family) |
| MSMEG_0861 Phosphatidylserine decarboxylase proenzyme (EC 4.1.1.65) [Cleaved into: Phosphatidylserine decarboxylase alpha chain; Phosphatidylserine decarboxylase beta chain] | MSMEG_1902 LysR-type transcriptional regulator (Transcriptional regulator) |
| MSMEG_0858 Cell division control protein Cdc48 (Conserved ATPase) (EC 3.6.4.6) | MSMEG_1903 Caib/baif family protein (L-carnitine dehydratase/bile acid-inducible protein F) |
| MSMEG_0457 Topoisomerase subunit TopoN (EC 5.99.1.3) (DNA gyrase subunit B-like protein MSMEG_0457/MSMEI_0444) | MSMEG_1904 Acyl-CoA dehydrogenase (Putative acyl-CoA dehydrogenase) |
| MSMEG_0456 Topoisomerase subunit TopoM (EC 5.99.1.3) | MSMEG_1905 Acyl-CoA dehydrogenase (EC 1.3.8.-) (Acyl-CoA dehydrogenase protein) |
| MSMEG_0459 Putative two-component system response regulator, LuxR family (Two-component system response regulator) | MSMEG_5215 Uncharacterized protein |
| MSMEG_0458 Two-component system sensor kinase | MSMEG_4901 Ribonuclease PH (RNase PH) (EC 2.7.7.56) (tRNA nucleotidyltransferase) |
| MSMEG_2143 SecB-like chaperone SmegB | MSMEG_3877 Precorrin-4 C11-methyltransferase (EC 2.1.1.133) |
| MSMEG_2145 Conserved domain protein | MSMEG_3875 Precorrin-6x reductase (EC 1.3.1.54) |
| MSMEG_2144 Uncharacterized protein | MSMEG_3950 Universal stress protein MSMEG_3950/MSMEI_3859 (USP MSMEG_3950) |
| MSMEG_3509 Uncharacterized protein | MSMEG_3878 Precorrin-6Y C5,15-methyltransferase (Decarboxylating) (EC 2.1.1.132) (Putative precorrin-6Y C5,15-methyltransferase CobL) (EC 2.1.1.132) |
| MSMEG_4899 Non-canonical purine NTP pyrophosphatase (EC 3.6.1.19) (Non-standard purine NTP pyrophosphatase) (Nucleoside-triphosphate diphosphatase) (Nucleoside-triphosphate pyrophosphatase) | MSMEG_3879 Short chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) |
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
X4XR.rep1
Bedaquiline.vs..DMSO.15min
M..smegmatis.Acid.NO.treated
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate1
KO_log.phase_rep2
Biological.replicate.2.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
MB100pruR.vs..MB100.rep.2
Biological.replicate.3.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
DglnR_without.vs..with.N_rep1
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.6.of.6..negative.control
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.2
X0.6..vs..50..air.saturation.Replicate.5
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.1.of.7..panaxydol.treatment
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
stphcy5plogcy3slide580norm
WT_log.phase_rep2
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Bedaquiline.vs..DMSO.60min
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
WT.vs..DglnR.N_starvation_rep1
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
Plogcy5plogcy3slide113norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
Biological.replicate.2.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments