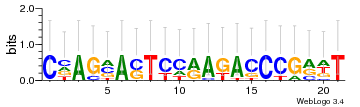
Module 542 Residual: 0.44
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.44 | 5.4e-30 | 0.028 | 77 | 31 | 20160204 |
| MSMEG_3642 Glycine dehydrogenase (decarboxylating) (EC 1.4.4.2) (Glycine cleavage system P-protein) (Glycine decarboxylase) (Glycine dehydrogenase (aminomethyl-transferring)) | MSMEG_6782 Uncharacterized protein |
| MSMEG_5049 Multifunctional 2-oxoglutarate metabolism enzyme (2-hydroxy-3-oxoadipate synthase) (HOA synthase) (HOAS) (EC 2.2.1.5) (2-oxoglutarate carboxy-lyase) (2-oxoglutarate decarboxylase) (Alpha-ketoglutarate decarboxylase) (KG decarboxylase) (KGD) (EC 4.1.1.71) (Alpha-ketoglutarate-glyoxylate carboligase) [Includes: 2-oxoglutarate dehydrogenase E1 component (ODH E1 component) (EC 1.2.4.2) (Alpha-ketoglutarate dehydrogenase E1 component) (KDH E1 component); Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex (EC 2.3.1.61) (2-oxoglutarate dehydrogenase complex E2 component) (ODH E2 component) (OGDC-E2) (Dihydrolipoamide succinyltransferase)] | MSMEG_6781 Uncharacterized protein |
| MSMEG_3059 Esterase | MSMEG_2048 Alcohol dehydrogenase (EC 1.1.1.1) |
| MSMEG_2310 | MSMEG_1030 Cyclohexanone monooxygenase (EC 1.14.13.22) (Monooxygenase) |
| MSMEG_1721 | MSMEG_6363 Cysteine desulfurase family protein (Putative aminotransferase/cysteine desulfurase) (EC 2.8.1.7) |
| MSMEG_3599 Periplasmic binding protein/LacI transcriptional regulator (Sugar-binding transcriptional regulator, LacI family protein) | MSMEG_2033 NADPH quinone oxidoreductase FadB4 (EC 1.6.5.5) (Oxidoreductase, zinc-binding dehydrogenase family protein) |
| MSMEG_1628 Oxidoreductase, short chain dehydrogenase/reductase family protein (Short-chain dehydrogenase/reductase SDR) | MSMEG_2034 Aryl-alcohol dehydrogenase (EC 1.1.1.90) |
| MSMEG_3202 Uncharacterized protein | MSMEG_1446 NTP pyrophosphohydrolase |
| MSMEG_4731 Acyl-CoA synthase (Fatty-acid-CoA ligase FadD23) | MSMEG_4257 Uncharacterized protein |
| MSMEG_2513 Uncharacterized protein | MSMEG_3609 Hydride transferase 1 |
| MSMEG_1626 Putative DNA-binding protein | MSMEG_5386 Putative Rhodanese-like domain protein (Rhodanese-like domain protein) |
| MSMEG_5032 Putative transcriptional regulator (Transcriptional regulator, HxlR family) | MSMEG_0385 Putative glycosyl transferase (EC 2.-.-.-) |
| MSMEG_5034 Uncharacterized protein | MSMEG_0241 Mycolic acid-containing lipids exporter MmpL11 |
| MSMEG_6852 Putative carboxylesterase/lipase | MSMEG_4259 DNA polymerase III, epsilon subunit |
| MSMEG_3060 Uncharacterized protein | MSMEG_2443 Uncharacterized protein |
| MSMEG_1736 Glycerol-3-phosphate dehydrogenase (EC 1.1.5.3) |
Bedaquiline.vs..DMSO.30min
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Plogcy5plogcy3slide110norm
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
KO_log.phase_rep2
KO_log.phase_rep1
DglnR_without.vs..with.N_rep1
Wild.type.rep1.y
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
stphcy5plogcy3slide487norm
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep2
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
X4dbfcy5plogcy3slide480norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.1.of.7..panaxydol.treatment
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
Biological.replicate.4.of.6..isoniazid.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
Comments
Log in to post comments