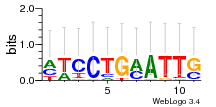
Module 562 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 4.7e-17 | 0.0000000037 | 83 | 26 | 20160204 |
| MSMEG_6428 Uncharacterized protein | MSMEG_4534 Putative conserved membrane protein (Uncharacterized protein) |
| MSMEG_4428 Uncharacterized protein | MSMEG_4535 Trehalase (EC 3.2.1.28) (Alpha,alpha-trehalase) (Alpha,alpha-trehalose glucohydrolase) |
| MSMEG_2759 Uncharacterized protein | MSMEG_2749 Uncharacterized protein |
| MSMEG_2419 Formamidopyrimidine-DNA glycosylase (Fapy-DNA glycosylase) (EC 3.2.2.23) (DNA-(apurinic or apyrimidinic site) lyase MutM) (AP lyase MutM) (EC 4.2.99.18) | MSMEG_2661 Lipoprotein (Uncharacterized protein) |
| MSMEG_2418 Ribonuclease 3 (EC 3.1.26.3) (Ribonuclease III) (RNase III) | MSMEG_2662 Uncharacterized protein |
| MSMEG_2417 Uncharacterized protein | MSMEG_1253 Uncharacterized protein |
| MSMEG_6197 Diaminopimelate decarboxylase (EC 4.1.1.20) | MSMEG_1254 DEAD/DEAH box helicase (DEAD/DEAH box helicase-like protein) |
| MSMEG_1265 Uncharacterized protein | MSMEG_2462 Carbon-monoxide dehydrogenase |
| MSMEG_2459 ABC transporter (Transporter ATPase) | MSMEG_2463 Carbon-monoxide dehydrogenase (EC 1.2.99.2) (Nicotine dehydrogenase chain A) |
| MSMEG_2458 Putative substrate-binding protein, ABC-type transporter (Uncharacterized protein) | MSMEG_2460 Transporter permease 1 |
| MSMEG_1128 Uncharacterized protein | MSMEG_2461 Binding-protein-dependent transport systems inner membrane component (Transporter permease 2, putative) |
| MSMEG_1129 D-amino acid dehydrogenase (EC 1.4.99.1) (D-amino-acid dehydrogenase) | MSMEG_3958 Glyoxalase/bleomycin resistance protein/dioxygenase |
| MSMEG_2484 Uncharacterized protein | MSMEG_2464 [2Fe-2S] binding domain protein |
Wild.type.rep1.y
KO_stationary.phase_rep1
KO_stationary.phase_rep2
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
X4XR.rep1
WT.vs..DamtR.without.N_rep2
Wild.type.rep2.x
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Bedaquiline.vs..DMSO.30min
Biological.replicate.1.of.6..negative.control
MB100pruR.vs..MB100..rep.3
Tet.CarD.vs.Control..Replicates.1..2..and.3.
dosR.WT_Replicate1
stphcy5plogcy3slide487norm
WT.vs..DglnR.N_starvation_dye.swap_rep1
Plogcy5plogcy3slide110norm
X3dbfcy5plogcy3slide601norm
DglnR_without.vs..with.N_rep1
MB100pruR.vs..MB100.rep.2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.2.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
DamtR.with.vs..DamtR.without.N_.rep2
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X0.6..vs..50..air.saturation.Replicate.5
Slow.vs..Fast.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Plogcy5plogcy3slide113norm
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.2.of.7..ethambutol.treatment
mc2155.vs.Dcrp1.replicate.4
X3dbfcy5plogcy3slide603norm
dcpA.knockout.rep1
Biological.replicate.2.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.5.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.4
Slow.vs..Fast.Replicate.5
WT.with.vs..without.N_dye.swap_rep2
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
dosR.WT_Replicate3
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
X4dbfcy5plogcy3slide901norm
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
Wild.type.rep1.x
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
rel.knockout.rep2
WT.vs..DglnR.N_starvation_dye.swap_rep2
WT.vs..DamtR.N.surplus_dyeswap_rep2
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments