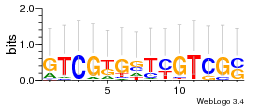
Module 591 Residual: 0.58
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.58 | 2.3e-21 | 0.0021 | 84 | 33 | 20160204 |
| MSMEG_3176 RarD protein | MSMEG_5527 Uncharacterized protein |
| MSMEG_3175 Pseudouridine synthase (EC 5.4.99.-) | MSMEG_3061 Primosomal protein N |
| MSMEG_3174 Lipoprotein signal peptidase (EC 3.4.23.36) (Prolipoprotein signal peptidase) (Signal peptidase II) | MSMEG_3060 Uncharacterized protein |
| MSMEG_3173 L-asparaginase (L-asparaginase AnsA) (EC 3.5.1.1) | MSMEG_5079 Uncharacterized protein |
| MSMEG_4110 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.-) | MSMEG_1819 N5-carboxyaminoimidazole ribonucleotide synthase (N5-CAIR synthase) (EC 6.3.4.18) (5-(carboxyamino)imidazole ribonucleotide synthetase) |
| MSMEG_3059 Esterase | MSMEG_1940 Hydrolase, alpha/beta fold family protein |
| MSMEG_4573 Competence protein (Uncharacterized protein) | MSMEG_1941 DNA helicase (EC 3.6.4.12) |
| MSMEG_5408 Prolipoprotein diacylglyceryl transferase (EC 2.4.99.-) | MSMEG_1943 ATP-dependent DNA helicase |
| MSMEG_1820 N5-carboxyaminoimidazole ribonucleotide mutase (N5-CAIR mutase) (EC 5.4.99.18) (5-(carboxyamino)imidazole ribonucleotide mutase) | MSMEG_1981 Uncharacterized protein |
| MSMEG_4407 Uncharacterized protein | MSMEG_0545 Transcriptional regulator, LuxR family protein (Transcriptional regulatory protein (LuxR-family)) |
| MSMEG_2040 Uncharacterized protein | MSMEG_2322 |
| MSMEG_0158 Formyl-CoA:oxalate CoA-transferase (FCOCT) (EC 2.8.3.16) (Formyl-coenzyme A transferase) | MSMEG_2039 Putative transcriptional regulator (Transcriptional regulator, AraC family) |
| MSMEG_6499 Uncharacterized protein | MSMEG_4316 Methylated-DNA--protein-cysteine methyltransferase (EC 2.1.1.63) (6-O-methylguanine-DNA methyltransferase) (O-6-methylguanine-DNA-alkyltransferase) |
| MSMEG_4321 Uncharacterized protein | MSMEG_4317 Uncharacterized protein |
| MSMEG_4320 Alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal allergen (EC 1.11.1.15) | MSMEG_4315 RNA polymerase sigma factor, sigma-70 family protein (RNA polymerase, sigma-24 subunit, ECF subfamily) |
| MSMEG_0259 Enoyl-CoA hydratase/isomerase | MSMEG_4090 Putative IclR family protein transcriptional regulator (Transcriptional regulator IclR) |
| MSMEG_5526 Peptidoglycan-binding LysM |
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
X4XR.rep1
Biological.replicate.3.of.7..falcarinol.treatment
dosR.WT_Replicate3
dosR.WT_Replicate1
Biological.replicate.5.of.6..negative.control
KO_log.phase_rep2
dosR.WT_Replicate4
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.1.of.6..isoniazid.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
Bedaquiline.vs..DMSO..45min
MB100pruR.vs..MB100.rep.2
Biological.replicate.6.of.7..falcarinol.treatment
DamtR.with.vs..DamtR.without.N_.rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.2.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
X0.6..vs..50..air.saturation.Replicate.5
Biological.replicate.6.of.7..ethambutol.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
stphcy5plogcy3slide487norm
stphcy3plogcy5slide581norm
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.7..falcarinol.treatment
X3dbfcy5plogcy3slide603norm
stphcy5plogcy3slide580norm
Biological.replicate.3.of.6..kanamycin.treatment
X2.5..vs..50..air.saturation.Replicate.5
X3dbfcy3plogcy5slide607norm
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide480norm
Biological.replicate.2.of.7..ethambutol.treatment
X2.5..vs..50..air.saturation.Replicate.3
Biological.replicate.2.of.6..negative.control
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
Biological.replicate.5.of.6..isoniazid.treatment
Bedaquiline.vs..DMSO.60min
Biological.replicate.4.of.7..falcarinol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Slow.vs..Fast.Replicate.2
Biological.replicate.7.of.7..panaxydol.treatment
Plogcy5plogcy3slide110norm
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_dyeswap_rep2
WT.vs..DglnR.N_starvation_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
Biological.replicate.1.of.6..negative.control
Biological.replicate.4.of.7..ethambutol.treatment
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Biological.replicate.6.of.7..panaxydol.treatment
Bedaquiline.vs..DMSO.120min
Comments
Log in to post comments