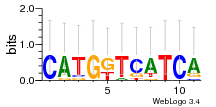
Module 617 Residual: 0.52
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.52 | 0.0063 | 270 | 77 | 37 | 20160204 |
| MSMEG_4791 | MSMEG_3256 Mucin-associated surface protein |
| MSMEG_5772 Cytospin-A, putative | MSMEG_6760 Activator of Hsp90 ATPase 1-like protein (Uncharacterized protein) |
| MSMEG_4628 Uncharacterized protein | MSMEG_0813 Uncharacterized protein |
| MSMEG_6614 Integral membrane protein (Metallophosphoesterase) | MSMEG_6762 Transcriptional regulator |
| MSMEG_5202 Conserved membrane protein | MSMEG_4002 Oxidoreductase, zinc-binding dehydrogenase family protein |
| MSMEG_5409 Uncharacterized protein | MSMEG_0455 Aldehyde Dehydrogenase (EC 1.2.1.-) (Aldehyde dehydrogenase) |
| MSMEG_0432 Uroporphyrinogen III synthase HEM4 (EC 4.2.1.75) (Uroporphyrinogen-III synthetase) | MSMEG_0187 Acetyltransferase |
| MSMEG_0431 Cobalamin (Vitamin B12) biosynthesis CbiX protein (EC 4.99.1.3) (Secreted protein) | MSMEG_4388 ABC transporter (ABC transporter ATP-binding protein) |
| MSMEG_0354 Uncharacterized protein | MSMEG_3557 Amino acid permease-associated region |
| MSMEG_6544 Transport-associated, putative | MSMEG_0522 Pp24 protein |
| MSMEG_3508 Hydrolase, alpha/beta hydrolase fold family protein | MSMEG_2267 Tetratricopeptide repeat domain protein |
| MSMEG_1760 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) | MSMEG_0189 Anhydro-N-acetylmuramic acid kinase (EC 2.7.1.170) (AnhMurNAc kinase) |
| MSMEG_6371 GntR-family protein regulator (GntR-family transcriptional regulator) (EC 2.6.1.39) | MSMEG_3856 Transcriptional regulator, CadC |
| MSMEG_0341 Putative oxidoreductase/luciferase-like protein (EC 1.5.99.11) (Uncharacterized protein) | MSMEG_2424 Signal recognition particle receptor FtsY (SRP receptor) |
| MSMEG_5822 Uncharacterized protein | MSMEG_3106 Quinone oxidoreductase (EC 1.6.5.5) |
| MSMEG_6859 Oxidoreductase | MSMEG_3666 C-5 sterol desaturase (EC 1.3.-.-) |
| MSMEG_5039 Siderophore-interacting protein | MSMEG_0127 Oxidoreductase, zinc-binding dehydrogenase family protein (Zinc-type alcohol dehydrogenase (E subunit) AdhE) (EC 1.1.1.1) |
| MSMEG_6538 3-oxoacyl-(Acyl-carrier-protein) reductase, putative (Short-chain dehydrogenase/reductase SDR) | MSMEG_4214 Glutamine amidotransferase |
| MSMEG_0955 Uncharacterized protein |
KO_stationary.phase_rep1
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DamtR.without.N_rep2
DamtR.with.vs..DamtR.without.N_.rep1
Mycobacterium.smegmatis.Spheroplast.induced
X4XR.rep1
rel.knockout.rep2
Biological.replicate.2.of.7..falcarinol.treatment
Biological.replicate.5.of.6..kanamycin.treatment
KO_log.phase_rep2
Plogcy5plogcy3slide110norm
Wild.type.rep1.y
WT.vs..DamtR.without.N_.dyeswap_rep1
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.1
WT_log.phase_rep2
Biological.replicate.4.of.6..kanamycin.treatment
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
Plogcy5plogcy3slide113norm
mc2155.vs.Dcrp1.replicate.1
mc2155.vs.Dcrp1.replicate.4
dcpA.knockout.rep2
dcpA.knockout.rep1
WT.vs..DamtR.N.surplus_rep1
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.4
X3dbfcy3plogcy5slide607norm
DglnR_without.vs..with.N_rep1
X2.5..vs..50..air.saturation.Replicate.1
X2.5..vs..50..air.saturation.Replicate.2
Biological.replicate.2.of.6..kanamycin.treatment
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
mc2155.vs.Dcrp1.replicate.3
stphcy5plogcy3slide487norm
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Bedaquiline.vs..DMSO.60min
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.6.of.7..ethambutol.treatment
Slow.vs..Fast.Replicate.3
X4dbfcy5plogcy3slide480norm
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
WT.vs..DamtR.N.surplus_.dyeswap_rep1
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
Biological.replicate.7.of.7..panaxydol.treatment
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M..smegmatis.Acid.NO.treated
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Comments
Log in to post comments