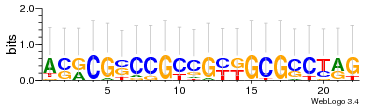
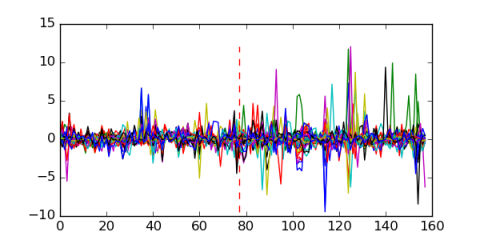
Module 628 Residual: 0.49
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.49 | 0.00019 | 0.00022 | 77 | 29 | 20160204 |
| MSMEG_4350 Dihydrodipicolinate reductase (EC 1.17.1.8) | MSMEG_3537 Uncharacterized protein |
| MSMEG_4351 Putative oxidoreductase YjgI (EC 1.-.-.-) (Short-chain dehydrogenase/reductase SDR) (EC 1.1.1.100) | MSMEG_4348 Acetolactate synthase large subunit (Thiamine pyrophosphate enzyme-like TPP-binding protein) (EC 2.2.1.6) |
| MSMEG_3357 Metal-dependent phosphohydrolase, HD subdomain (Metal-dependent phosphohydrolase, HD subdomain protein) | MSMEG_5216 Glyoxalase family protein (Glyoxalase/bleomycin resistance protein/dioxygenase) |
| MSMEG_0684 Aldehyde oxidase and xanthine dehydrogenase molybdopterin binding protein (EC 1.17.1.4) (Aldehyde oxidase and xanthine dehydrogenase, molybdopterin binding) | MSMEG_0367 O-demethylpuromycin-O-methyltransferase (EC 2.1.1.38) |
| MSMEG_0685 Molybdopterin dehydrogenase, FAD-binding protein (EC 1.17.1.4) (Oxidoreductase, molybdopterin-binding subunit) | MSMEG_5700 Secreted protein (Transglycosylase domain protein) |
| MSMEG_0686 (2Fe-2S)-binding protein (EC 1.2.99.2) (Oxidoreductase) | MSMEG_0370 Uncharacterized protein |
| MSMEG_3536 Major facilitator superfamily MFS_1 (Sugar transport protein) | MSMEG_0369 Uncharacterized protein |
| MSMEG_1621 Pyrimidine-specific ribonucleoside hydrolase RihA (EC 3.2.-.-) | MSMEG_0368 Conserved hypothetical alanine and proline rich protein (Uncharacterized protein) |
| MSMEG_0451 Oxidoreductase, FAD-linked (Uncharacterized protein) | MSMEG_0465 Transcriptional regulator, TetR family (Transcriptional regulator, TetR family protein) |
| MSMEG_0235 Probable conserved membrane protein | MSMEG_4596 Uncharacterized protein |
| MSMEG_0234 Metallopeptidase (PgPepO oligopeptidase Metallo peptidase MEROPS family M13) (EC 3.4.24.-) | MSMEG_0089 Chromosome condensation protein (Pirin-like protein) |
| MSMEG_0236 Putative conserved transmembrane protein | MSMEG_1569 Uncharacterized protein |
| MSMEG_4246 Conserved integral membrane protein (Probable conserved integral membrane protein) | MSMEG_0123 Uncharacterized protein |
| MSMEG_4247 Polyprenol-phosphate-mannose-dependent alpha-(1-2)-phosphatidylinositol mannoside mannosyltransferase (EC 2.4.1.-) (Alpha-D-mannose-alpha-(1-2)-mannosyltransferase) (Alpha-mannosyltransferase) (Alpha-ManT) (PPM-dependent mannosyltransferase) (Polyprenol-phosphate-mannose alpha-mannosyltransferase) (PPM alpha-mannosyltransferase) | MSMEG_0122 Putative periplasmic solute-binding protein (Uncharacterized protein UPF0065) |
| MSMEG_6389 Integral membrane indolylacetylinositol arabinosyltransferase EmbB (EC 2.4.2.34) (Probable arabinosyltransferase A) (EC 2.4.2.-) |
KO_stationary.phase_rep1
Wild.type.rep2.y
Bedaquiline.vs..DMSO.15min
Mycobacterium.smegmatis.Spheroplast.induced
Biological.replicate.2.of.6..isoniazid.treatment
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.1.of.6..negative.control
dosR.WT_Replicate1
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
MB100pruR.vs..MB100.rep.1
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.6.of.6..kanamycin.treatment
Biological.replicate.3.of.6..kanamycin.treatment
Wild.type.rep3
Bedaquiline.vs..DMSO..45min
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
M..smegmatis.Acid.NO.treated
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
WT.vs..DamtR.without.N_.dyeswap_rep1
WT_log.phase_rep2
X0.6..vs..50..air.saturation.Replicate.2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
Slow.vs..Fast.Replicate.3
X3dbfcy3plogcy5slide607norm
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
dcpA.knockout.rep2
Biological.replicate.2.of.7..falcarinol.treatment
stphcy5plogcy3slide580norm
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
WT_log.phase_rep1
Biological.replicate.4.of.7..panaxydol.treatment
X2.5..vs..50..air.saturation.Replicate.1
Biological.replicate.2.of.7..panaxydol.treatment
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
Biological.replicate.4.of.6..isoniazid.treatment
X4XR.rep1
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
Biological.replicate.5.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.2
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
DglnR_without.vs..with.N_dyeswap_rep2
X2XR.rep1
Wild.type.rep1.y
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Plogcy5plogcy3slide113norm
BatchCulture_LBTbroth_StationaryPhase_Replicate3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.3.of.6..negative.control
X4dbfcy3plogcy5slide903norm
Comments
Log in to post comments