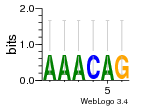
Module 642 Residual: 0.42
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.42 | 110 | 8000 | 83 | 28 | 20160204 |
| MSMEG_4118 Acyl-CoA dehydrogenase (EC 1.3.99.-) (Isovaleryl-CoA dehydrogenase) (EC 1.3.8.4) | MSMEG_1470 50S ribosomal protein L6 |
| MSMEG_4119 3-hydroxybutyryl-CoA dehydratase (EC 4.2.1.55) (Enoyl-CoA hydratase) (EC 4.2.1.17) | MSMEG_1471 50S ribosomal protein L18 |
| MSMEG_1521 30S ribosomal protein S13 | MSMEG_1472 30S ribosomal protein S5 |
| MSMEG_4111 Alpha-methylacyl-CoA racemase (L-carnitine dehydratase/bile acid-inducible protein F) (EC 5.1.99.4) | MSMEG_1473 50S ribosomal protein L30 |
| MSMEG_4112 Cyclohexanecarboxylate-CoA ligase | MSMEG_6897 30S ribosomal protein S6 |
| MSMEG_4113 Acyl-CoA dehydrogenase (Uncharacterized protein) | MSMEG_4120 CAIB/BAIF family protein |
| MSMEG_4114 1,4-Dihydroxy-2-naphthoate synthase (EC 4.1.3.36) (Naphthoate synthase) (EC 4.1.3.36) | MSMEG_1467 50S ribosomal protein L5 |
| MSMEG_4115 3-hydroxybutyryl-CoA dehydrogenase (EC 1.1.1.157) (3-hydroxybutyryl-CoA dehydrogenase, 3-hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.-) | MSMEG_1466 50S ribosomal protein L24 |
| MSMEG_4116 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.-) (3-hydroxybutyryl-CoA dehydrogenase) (EC 1.1.1.157) | MSMEG_1465 50S ribosomal protein L14 |
| MSMEG_4117 2-deoxy-D-gluconate 3-dehydrogenase (EC 1.1.1.125) | MSMEG_1522 30S ribosomal protein S11 |
| MSMEG_1439 50S ribosomal protein L2 | MSMEG_1523 30S ribosomal protein S4 |
| MSMEG_1435 30S ribosomal protein S10 | MSMEG_1469 30S ribosomal protein S8 |
| MSMEG_1436 50S ribosomal protein L3 | MSMEG_1468 30S ribosomal protein S14 type Z |
| MSMEG_1474 50S ribosomal protein L15 | MSMEG_1443 50S ribosomal protein L16 |
KO_stationary.phase_rep2
WT.vs..DglnR.N_starvation_rep2
X4XR.rep1
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Bedaquiline.vs..DMSO.30min
X2XR.rep1
Biological.replicate.4.of.6..negative.control
dosR.WT_Replicate2
dosR.WT_Replicate1
KO_log.phase_rep2
KO_log.phase_rep1
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep2
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.7.of.7..ethambutol.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
WT.with.vs..without.N_dye.swap_rep1
Biological.replicate.3.of.7..falcarinol.treatment
Wild.type.rep3
Biological.replicate.1.of.7..falcarinol.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Biological.replicate.1.of.6..negative.control
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
Biological.replicate.5.of.7..falcarinol.treatment
DglnR_without.vs..with.N_rep1
Biological.replicate.4.of.6..isoniazid.treatment
X0.6..vs..50..air.saturation.Replicate.4
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.1
X3dbfcy3plogcy5slide607norm
Biological.replicate.3.of.6..negative.control
X4dbfcy5plogcy3slide901norm
Biological.replicate.5.of.7..panaxydol.treatment
Biological.replicate.2.of.6..isoniazid.treatment
stphcy5plogcy3slide580norm
X3dbfcy5plogcy3slide601norm
X2.5..vs..50..air.saturation.Replicate.5
stphcy3plogcy5slide581norm
Biological.replicate.4.of.7..panaxydol.treatment
X4dbfcy5plogcy3slide480norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Biological.replicate.5.of.7..ethambutol.treatment
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.5.of.6..isoniazid.treatment
WT.vs..DamtR.N.surplus_dyeswap_rep2
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
BatchCulture_LBTbroth_ExponentialPhase_Replicate3
X3dbfcy5plogcy3slide603norm
Biological.replicate.5.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.vs..DglnR.N_starvation_rep1
WT.with.vs..without.N_rep1
Log.phase.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
X2XR.rep2
DglnR_without.vs..with.N_dyeswap_rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate4
Sub.MIC.INH
BatchCulture_LBTbroth_StationaryPhase_Replicate2
BatchCulture_LBTbroth_StationaryPhase_Replicate3
BatchCulture_LBTbroth_StationaryPhase_Replicate1
Biological.replicate.2.of.6..negative.control
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep1
Biological.replicate.2.of.7..ethambutol.treatment
Biological.replicate.6.of.7..panaxydol.treatment
Comments
Log in to post comments