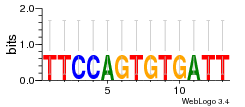
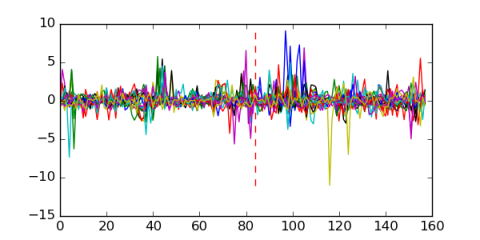
Module 656 Residual: 0.57
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.57 | 2.4e-20 | 5.2 | 84 | 20 | 20160204 |
| MSMEG_5974 Uncharacterized protein | MSMEG_6113 D-alanyl-D-alanine carboxypeptidase DacB1 (EC 3.4.16.4) (D-alanyl-D-alanine carboxypeptidase/D-alanyl-D-alanine-endopeptidase) (EC 3.4.16.4) |
| MSMEG_5645 Uncharacterized protein | MSMEG_4141 Uncharacterized protein |
| MSMEG_2763 Uncharacterized protein | MSMEG_6111 tRNA(Ile)-lysidine synthase (EC 6.3.4.19) (tRNA(Ile)-2-lysyl-cytidine synthase) (tRNA(Ile)-lysidine synthetase) |
| MSMEG_5300 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) (Short-chain type dehydrogenase/reductase) (EC 1.-.-.-) | MSMEG_2761 Putative secreted alanine rich protein (Uncharacterized protein) |
| MSMEG_4527 Ferredoxin sulfite reductase (EC 1.8.7.1) (Ferredoxin-nitrite reductase) (EC 1.-.-.-) | MSMEG_6114 Inorganic pyrophosphatase (EC 3.6.1.1) |
| MSMEG_5563 AraC-family protein transcriptional regulator (AraC-family transcriptional regulator) | MSMEG_5301 Transcriptional regulator (Transcriptional regulator, TetR family) |
| MSMEG_0388 Macrocin-O-methyltransferase (EC 2.1.1.101) | MSMEG_4529 Secreted protein |
| MSMEG_5564 NAD(P)H dehydrogenase, quinone family protein | MSMEG_4528 Phosphoadenosine phosphosulfate reductase (EC 1.8.4.8) |
| MSMEG_0389 Gtf1 (Putative glycosyl transferase) (EC 2.-.-.-) (Uncharacterized protein) | MSMEG_5644 Membrane protein |
| MSMEG_6112 Uncharacterized protein | MSMEG_6823 Short-chain dehydrogenase/reductase SDR (EC 1.1.1.100) |
Wild.type.rep1.y
KO_stationary.phase_rep1
Bedaquiline.vs..DMSO.30min
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Stationary.cultures.induced.to.express.I.SceI.for.45.minutes..T.45...ATc.
WT.vs..DamtR.without.N_rep2
M..smegmatis.Acid.NO.treated
Biological.replicate.4.of.6..negative.control
Wild.type.rep2.x
Biological.replicate.4.of.7..ethambutol.treatment
Wild.type.rep3
Biological.replicate.3.of.7..ethambutol.treatment
MB100pruR.vs..MB100..rep.3
dosR.WT_Replicate2
Biological.replicate.2.of.6..kanamycin.treatment
Wild.type.rep2.y
Plogcy5plogcy3slide110norm
Biological.replicate.7.of.7..falcarinol.treatment
MB100pruR.vs..MB100.rep.4
Biological.replicate.3.of.7..panaxydol.treatment
Biological.replicate.6.of.6..negative.control
DglnR_without.vs..with.N_rep2
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..isoniazid.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.3.of.7..falcarinol.treatment
rel.knockout.rep1
Bedaquiline.vs..DMSO..45min
Biological.replicate.5.of.6..isoniazid.treatment
Biological.replicate.6.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.1
X3dbfcy5plogcy3slide601norm
X0.6..vs..50..air.saturation.Replicate.3
X0.6..vs..50..air.saturation.Replicate.5
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
Plogcy5plogcy3slide113norm
Biological.replicate.5.of.6..kanamycin.treatment
X3dbfcy3plogcy5slide607norm
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
X2.5..vs..50..air.saturation.Replicate.4
stphcy3plogcy5slide581norm
WT_log.phase_rep1
X3dbfcy5plogcy3slide603norm
X2.5..vs..50..air.saturation.Replicate.2
X2.5..vs..50..air.saturation.Replicate.3
WT_stationary.phase_rep2
WT_stationary.phase_rep1
Biological.replicate.6.of.6..isoniazid.treatment
X4XR.rep2
DamtR.with.vs..DamtR.without.N_dyeswap_.rep1
Slow.vs..Fast.Replicate.4
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
dosR.WT_Replicate3
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
BatchCulture_LBTbroth_ExponentialPhase_Replicate1
Biological.replicate.3.of.6..kanamycin.treatment
mc2155.pMind..vs.mc2155.pMind.6189.replicate.3
BatchCulture_LBTbroth_ExponentialPhase_Replicate4
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
Biological.replicate.1.of.7..panaxydol.treatment
Log.phase.cultures.without.induction.of.I.SceI.expression..T.0...Atc.
WT.vs..DglnR.N_starvation_rep2
DglnR_without.vs..with.N_dyeswap_rep2
Biological.replicate.1.of.6..negative.control
DglnR_without.vs..with.N_dyeswap_rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
BatchCulture_LBTbroth_StationaryPhase_Replicate2
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep4
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Biological.replicate.2.of.7..ethambutol.treatment
WT.vs..DglnR.N_starvation_dye.swap_rep1
Comments
Log in to post comments