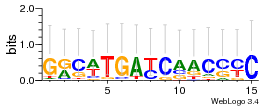
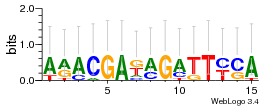
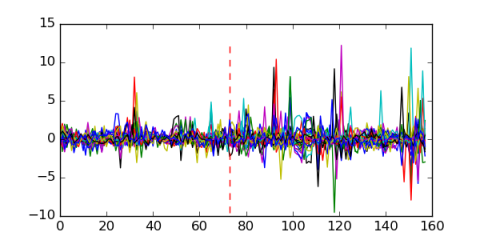
Module 667 Residual: 0.46
Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155)
| Residual | Motif 1 e-value | Motif 2 e-value | Conditions | Genes | version |
|---|---|---|---|---|---|
| 0.46 | 0.0025 | 6.9 | 73 | 29 | 20160204 |
| MSMEG_5209 Hydrolase, alpha/beta fold family protein (Putative peroxidase (Non-heme peroxidase), BpoB, alpha/beta hydrolase family) (EC 1.11.1.10) | MSMEG_2006 2,3-dihydroxybiphenyl 1,2-dioxygenase |
| MSMEG_5464 Integral membrane transport protein (Major facilitator superfamily MFS_1) | MSMEG_0019 Amino acid adenylation (Peptide synthetase) |
| MSMEG_5463 6-phosphogluconate dehydrogenase NAD-binding protein (EC 1.1.1.31) (6-phosphogluconate dehydrogenase, NAD-binding) | MSMEG_1120 Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase |
| MSMEG_6849 LysR-family protein transcriptional regulator (LysR-family transcriptional regulator) | MSMEG_4838 Uncharacterized protein |
| MSMEG_3424 S-adenosyl-L-methionine-dependent methyltransferase (EC 2.1.1.-) | MSMEG_5211 Aminotransferase class-III |
| MSMEG_1642 ABC transporter, ATP-binding protein | MSMEG_4722 Short-chain dehydrogenase (Short-chain dehydrogenase/reductase SDR) |
| MSMEG_0472 Putative lipoprotein | MSMEG_4721 Permease of the major facilitator superfamily protein |
| MSMEG_3039 Monooxygenase (Monooxygenase FAD-binding protein) | MSMEG_5410 Radical SAM (Radical SAM domain protein) |
| MSMEG_3465 Acyl-CoA ligase FadD31 (Acyl-CoA synthase) | MSMEG_0663 TetR-family protein transcriptional regulator (TetR-family transcriptional regulator) |
| MSMEG_1990 Conserved hypothetical cytosolic protein (Uncharacterized protein) | MSMEG_5365 RNA polymerase sigma-70 factor |
| MSMEG_1108 Hydrolase, alpha/beta fold family protein | MSMEG_1196 SNF2 domain protein (Uncharacterized protein) |
| MSMEG_1663 Transcriptional regulator, AsnC family protein | MSMEG_5845 Putative C4-dicarboxylate transporter (Uncharacterized protein) |
| MSMEG_1662 Taurine-pyruvate aminotransferase | MSMEG_1685 Uncharacterized protein |
| MSMEG_1665 Aldehyde dehydrogenase (NAD) family protein (EC 1.2.1.-) | MSMEG_1316 Peptide chain release factor 3 |
| MSMEG_1664 Uncharacterized protein |
Wild.type.rep1.y
KO_stationary.phase_rep2
Biological.replicate.5.of.7..falcarinol.treatment
WT.vs..DamtR.without.N_rep1
Wild.type.rep2.y
WT.vs..DamtR.without.N_rep2
Mycobacterium.smegmatis.Spheroplast.induced
dosR.WT_Replicate4
Biological.replicate.3.of.7..ethambutol.treatment
Biological.replicate.4.of.6..kanamycin.treatment
KO_log.phase_rep1
WT.vs..DamtR.N.surplus_dyeswap_rep2
Wild.type.rep1.x
Biological.replicate.7.of.7..falcarinol.treatment
Biological.replicate.6.of.6..negative.control
WT.vs..DamtR.without.N_.dyeswap_rep1
WT.vs..DamtR.without.N_.dyeswap_rep2
Biological.replicate.1.of.6..kanamycin.treatment
Biological.replicate.7.of.7..ethambutol.treatment
Biological.replicate.1.of.7..ethambutol.treatment
Biological.replicate.6.of.6..kanamycin.treatment
rel.knockout.rep2
X4XR.rep1
Biological.replicate.3.of.7..falcarinol.treatment
Bedaquiline.vs..DMSO.15min
DamtR.with.vs..DamtR.without.N_.rep1
Biological.replicate.2.of.7..panaxydol.treatment
Biological.replicate.3.of.6..isoniazid.treatment
DamtR.with.vs..DamtR.without.N_.rep2
X0.6..vs..50..air.saturation.Replicate.3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep1
WT.vs..DglnR.N_starvation_dye.swap_rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep3
M.smegmatis_mc2_155_dMSMEG_0166_study_mutant_rep2
mc2155.vs.Dcrp1.replicate.2
mc2155.vs.Dcrp1.replicate.3
mc2155.vs.Dcrp1.replicate.1
Biological.replicate.4.of.6..negative.control
mc2155.vs.Dcrp1.replicate.4
WT.vs..DamtR.N.surplus_rep2
dcpA.knockout.rep1
Biological.replicate.2.of.7..falcarinol.treatment
Plogcy5plogcy3slide111norm
Biological.replicate.2.of.6..isoniazid.treatment
X2.5..vs..50..air.saturation.Replicate.4
X2.5..vs..50..air.saturation.Replicate.5
WT_log.phase_rep1
X2.5..vs..50..air.saturation.Replicate.3
WT.vs..DglnR.N_starvation_dye.swap_rep1
Biological.replicate.6.of.6..isoniazid.treatment
Biological.replicate.5.of.7..ethambutol.treatment
Tet.CarD.vs.Control..Replicates.1..2..and.3.
Slow.vs..Fast.Replicate.5
Stationary.cultures.control.without.induction.of.I.SceI.expression..T.0...Atc.
Biological.replicate.4.of.7..falcarinol.treatment
Slow.vs..Fast.Replicate.1
Biological.replicate.6.of.7..ethambutol.treatment
Biological.replicate.7.of.7..panaxydol.treatment
BatchCulture_LBTbroth_ExponentialPhase_Replicate2
mc2155.pMind..vs.mc2155.pMind.6189.replicate.4
Biological.replicate.5.of.6..kanamycin.treatment
X4dbfcy5plogcy3slide901norm
WT.vs..DamtR.N.surplus_.dyeswap_rep1
WT.with.vs..without.N_rep2
WT.vs..DglnR.N_starvation_rep2
Wild.type.rep2.x
Biological.replicate.1.of.6..negative.control
mc2155.pMind..vs.mc2155.pMind.6189.replicate.1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep1
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep3
JR121..pJR29.VapC..vs..JR121..pJR230..VapBC..rep2
M.smegmatis_mc2_155_dMSMEG_0166_study_wild.type_rep2
Stationary.phase.cultures.induced.to.express.I.SceI.for.15.minutes..T.15...ATc.
Comments
Log in to post comments