41979 PRK06116Thalassiosira pseudonana
| Chromosome | Product | Transcript Start | End | Strand | Short Name | |
|---|---|---|---|---|---|---|
| 41979 | chr_11a | PRK06116 | 308948 | 310853 | - | PRK06116 |
| NCBI ID | Ensembl Genomes exon ID |
|---|---|
| 7443289 | Thaps41979.1, Thaps41979.2, Thaps41979.3 |
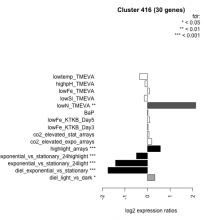
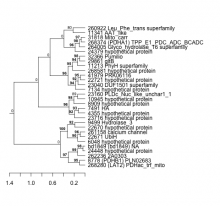
| Expression Profile | Conditional Changes | Cluster Dendrogram | Discovered Potential cis-Regulatory Motifs |
|---|---|---|---|
Thaps_hclust_0416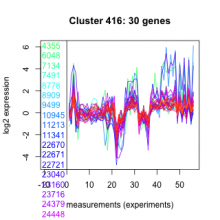 |
 |
 |
  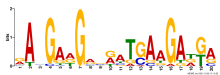 |
| Normalized Mean Residue | Discovered Potential cis-Regulatory Motifs | |
|---|---|---|
|
Thaps_bicluster_0150 |
0.38 |
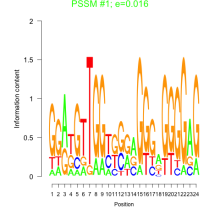 0.016 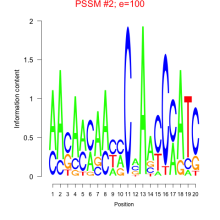 100 |

Add comment