32522 rad3Thalassiosira pseudonana
| Chromosome | Product | Transcript Start | End | Strand | Short Name | |
|---|---|---|---|---|---|---|
| 32522 | chr_3 | rad3 | 1577350 | 1580602 | + | rad3 |
| NCBI ID | Ensembl Genomes exon ID |
|---|---|
| 7449515 | Thaps32522.1, Thaps32522.4, Thaps32522.6, Thaps32522.2, Thaps32522.3, Thaps32522.5 |
| Expression Profile | Conditional Changes | Cluster Dendrogram | Discovered Potential cis-Regulatory Motifs |
|---|---|---|---|
Thaps_hclust_0261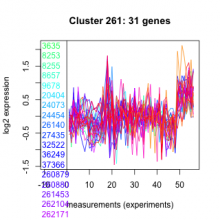 |
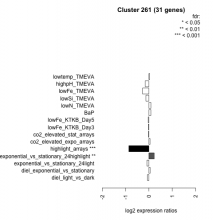 |
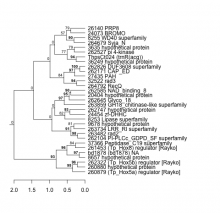 |
   |
| Normalized Mean Residue | Discovered Potential cis-Regulatory Motifs | |
|---|---|---|
|
Thaps_bicluster_0022 |
0.44 |
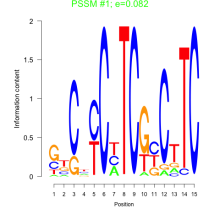 0.082 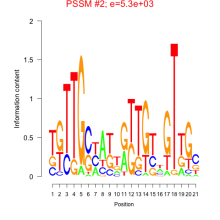 5300 |
| T. pseudonana | P. tricornutum | P. tricornutum DiatomCyc | F. cylindrus | Pseudo-nitzschia multiseries | E. huxleyi | C. reinhardtii | A. thaliana | P. sojae |
|---|---|---|---|---|---|---|---|---|
| Not available | PHATRDRAFT_24915 | PHATRDRAFT_24915 | 136107 | 200477 | 198134 | Cre02.g089608.t1.2 | AT1G79950.1 | 485995 |
| KEGG description | KEGG Pathway |
|---|---|
| Not available | Not available |

Add comment