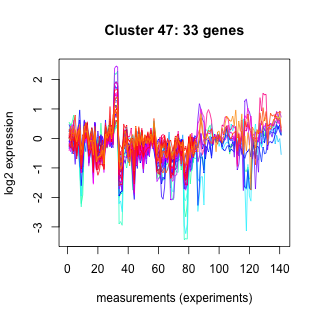
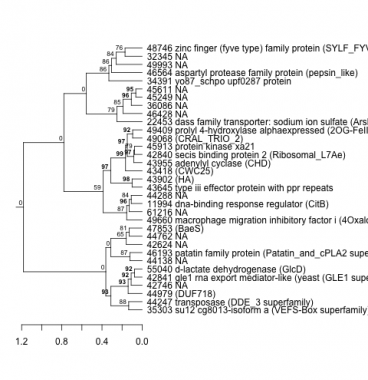
Phatr_hclust_0047 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0000160 | two-component signal transduction system (phosphorelay) | 0.00251588 | 1 | Phatr_hclust_0047 |
| GO:0005978 | glycogen biosynthesis | 0.00591934 | 1 | Phatr_hclust_0047 |
| GO:0016310 | phosphorylation | 0.0407661 | 1 | Phatr_hclust_0047 |
| GO:0006813 | potassium ion transport | 0.0577662 | 1 | Phatr_hclust_0047 |
| GO:0019538 | protein metabolism | 0.101759 | 1 | Phatr_hclust_0047 |
| GO:0007242 | intracellular signaling cascade | 0.117765 | 1 | Phatr_hclust_0047 |
| GO:0007165 | signal transduction | 0.130903 | 1 | Phatr_hclust_0047 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.134605 | 1 | Phatr_hclust_0047 |
| GO:0006468 | protein amino acid phosphorylation | 0.406444 | 1 | Phatr_hclust_0047 |
| GO:0006508 | proteolysis and peptidolysis | 0.495352 | 1 | Phatr_hclust_0047 |
|
PHATRDRAFT_35303 : su12 cg8013-isoform a (VEFS-Box superfamily) |
PHATRDRAFT_43418 : (CWC25) |
||
|
PHATRDRAFT_44247 : transposase (DDE_3 superfamily) |
PHATRDRAFT_43955 : adenylyl cyclase (CHD) |
||
|
PHATRDRAFT_44979 : (DUF718) |
PHATRDRAFT_47853 : (BaeS) |
PHATRDRAFT_42840 : secis binding protein 2 (Ribosomal_L7Ae) |
|
|
PHATRDRAFT_49660 : macrophage migration inhibitory factor i (4Oxalocrotonate_Tautomerase superfamily) |
PHATRDRAFT_45913 : protein kinase xa21 |
PHATRDRAFT_34391 : yo87_schpo upf0287 protein |
|
|
PHATRDRAFT_42841 : gle1 rna export mediator-like (yeast (GLE1 superfamily) |
PHATRDRAFT_11994 : dna-binding response regulator (CitB) |
PHATRDRAFT_49068 : (CRAL_TRIO_2) |
PHATRDRAFT_46564 : aspartyl protease family protein (pepsin_like) |
|
PHATRDRAFT_55040 : d-lactate dehydrogenase (GlcD) |
PHATRDRAFT_49409 : prolyl 4-hydroxylase alphaexpressed (2OG-FeII_Oxy_3 superfamily) |
||
|
PHATRDRAFT_43645 : type iii effector protein with ppr repeats |
PHATRDRAFT_22453 : dass family transporter: sodium ion sulfate (ArsB_NhaD_permease superfamily) |
||
|
PHATRDRAFT_46193 : patatin family protein (Patatin_and_cPLA2 superfamily) |
PHATRDRAFT_43902 : (HA) |
PHATRDRAFT_48746 : zinc finger (fyve type) family protein (SYLF_FYVE) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | -0.142 | 0.47642 |
| Copper_SH | Copper_SH | 0.120 | 0.165895 |
| Green_vs_Red_24h | Green_vs_Red_24h | 0.097 | 0.341266 |
| Silver_SH | Silver_SH | 0.280 | 0.00107527 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.102 | 0.0818182 |
| BlueLight_24h | BlueLight_24h | -0.281 | 0.00892857 |
| GreenLight_0.5h | GreenLight_0.5h | -0.970 | 0.000458716 |
| RedLight_6h | RedLight_6h | -0.566 | 0.00131579 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |
| GreenLight_24h | GreenLight_24h | -0.351 | 0.000961538 |
| light_6hr | light_6hr | -0.253 | 0.173785 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.060 | 0.767173 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | 0.013 | 0.908562 |
| Ammonia_SH | Ammonia_SH | -0.326 | 0.14816 |
| Simazine_SH | Simazine_SH | 0.467 | 0.000423729 |
| Oil_SH | Oil_SH | -0.173 | 0.0525463 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | 0.319 | 0.00604839 |
| Cadmium_SH | Cadmium_SH | 0.030 | 0.773152 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | 0.167 | 0.195029 |
| highlight_0to6h | highlight_0to6h | -0.127 | 0.352724 |
| BlueLight_0.5h | BlueLight_0.5h | -1.318 | 0.000869565 |
| Dispersant_SH | Dispersant_SH | 0.075 | 0.404493 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.184 | 0.0688571 |
| light_15.5hr | light_15.5hr | -0.329 | 0.096114 |
| light_10.5hr | light_10.5hr | -0.219 | 0.334281 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.227 | 0.0212418 |
| Si_free | Si_free | -0.029 | 0.785958 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | -1.181 | 0.000396825 |
| Mixture_SH | Mixture_SH | -0.391 | 0.101186 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.195 | 0.245578 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.244 | 0.229241 |
| BlueLight_6h | BlueLight_6h | -0.371 | 0.00643939 |
| highlight_12to48h | highlight_12to48h | -0.419 | 0.00487805 |
| RedLight_24h | RedLight_24h | -0.448 | 0.000943396 |
| Dark_treated | Dark_treated | 1.133 | 0.011747 |
| GreenLight_6h | GreenLight_6h | -0.385 | 0.0091195 |
| Cadmium_0.12mg | Cadmium_0.12mg | 0.046 | 0.18037 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | 0.070 | 0.273429 |
| RedLight_0.5h | RedLight_0.5h | -0.703 | 0.0173469 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.181 | 0.267219 |
| Re-illuminated_24h | Re-illuminated_24h | -0.376 | 0.000657895 |