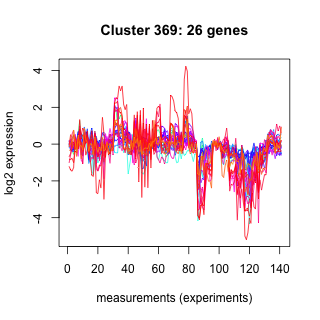
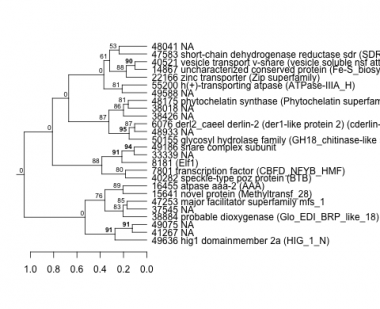
Phatr_hclust_0369 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0005975 | carbohydrate metabolism | 0.104035 | 1 | Phatr_hclust_0369 |
| GO:0006030 | chitin metabolism | 0.00172882 | 1 | Phatr_hclust_0369 |
| GO:0006886 | intracellular protein transport | 0.0671851 | 1 | Phatr_hclust_0369 |
| GO:0008152 | metabolism | 0.0865285 | 1 | Phatr_hclust_0369 |
| GO:0030001 | metal ion transport | 0.0273554 | 1 | Phatr_hclust_0369 |
| GO:0006810 | transport | 0.287164 | 1 | Phatr_hclust_0369 |
|
PHATRDRAFT_49636 : hig1 domainmember 2a (HIG_1_N) |
PHATRDRAFT_16455 : atpase aaa-2 (AAA) |
PHATRDRAFT_55200 : h(+)-transporting atpase (ATPase-IIIA_H) |
|
|
PHATRDRAFT_40282 : speckle-type poz protein (BTB) |
PHATRDRAFT_6076 : derl2_caeel derlin-2 (der1-like protein 2) (cderlin-2) (DER1 superfamily) |
PHATRDRAFT_22166 : zinc transporter (Zip superfamily) |
|
|
PHATRDRAFT_7801 : transcription factor (CBFD_NFYB_HMF) |
PHATRDRAFT_14867 : uncharacterized conserved protein (Fe-S_biosyn superfamily) |
||
|
PHATRDRAFT_38884 : probable dioxygenase (Glo_EDI_BRP_like_18) |
PHATRDRAFT_8181 : (Elf1) |
PHATRDRAFT_40521 : vesicle transport v-snare (vesicle soluble nsf attachment protein receptor) (V-SNARE) |
|
|
PHATRDRAFT_48175 : phytochelatin synthase (Phytochelatin superfamily) |
PHATRDRAFT_47583 : short-chain dehydrogenase reductase sdr (SDR_c) |
||
|
PHATRDRAFT_47253 : major facilitator superfamily mfs_1 |
PHATRDRAFT_49186 : snare complex subunit |
||
|
PHATRDRAFT_15641 : novel protein (Methyltransf_28) |
PHATRDRAFT_50155 : glycosyl hydrolase family (GH18_chitinase-like superfamily) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.459 | 0.0057554 |
| Copper_SH | Copper_SH | -0.356 | 0.00169492 |
| Green_vs_Red_24h | Green_vs_Red_24h | 0.061 | 0.612158 |
| Silver_SH | Silver_SH | 0.073 | 0.405256 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.022 | 0.825754 |
| BlueLight_24h | BlueLight_24h | -0.095 | 0.701384 |
| GreenLight_0.5h | GreenLight_0.5h | 0.390 | 0.248308 |
| RedLight_6h | RedLight_6h | 0.095 | 0.673951 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |
| GreenLight_24h | GreenLight_24h | -0.073 | 0.698722 |
| light_6hr | light_6hr | 0.302 | 0.144424 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | 0.257 | 0.171285 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | 0.037 | 0.77145 |
| Ammonia_SH | Ammonia_SH | -1.675 | 0.000344828 |
| Simazine_SH | Simazine_SH | -0.038 | 0.730407 |
| Oil_SH | Oil_SH | -1.105 | 0.000666667 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.637 | 0.000609756 |
| Cadmium_SH | Cadmium_SH | -0.739 | 0.000666667 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | 0.039 | 0.822055 |
| highlight_0to6h | highlight_0to6h | 0.076 | 0.720646 |
| BlueLight_0.5h | BlueLight_0.5h | 0.750 | 0.0665025 |
| Dispersant_SH | Dispersant_SH | -0.803 | 0.000909091 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.083 | 0.497959 |
| light_15.5hr | light_15.5hr | -0.274 | 0.235976 |
| light_10.5hr | light_10.5hr | 0.103 | 0.699634 |
| Dispersed_oil_SH | Dispersed_oil_SH | -1.144 | 0.000675676 |
| Si_free | Si_free | -0.083 | 0.473963 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 0.683 | 0.0618182 |
| Mixture_SH | Mixture_SH | -1.930 | 0.000357143 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.137 | 0.501288 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.250 | 0.282278 |
| BlueLight_6h | BlueLight_6h | 0.232 | 0.120722 |
| highlight_12to48h | highlight_12to48h | -0.266 | 0.132533 |
| RedLight_24h | RedLight_24h | -0.134 | 0.388793 |
| Dark_treated | Dark_treated | 1.122 | 0.0255435 |
| GreenLight_6h | GreenLight_6h | 0.195 | 0.2315 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.016 | 0.731063 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.022 | 0.816842 |
| RedLight_0.5h | RedLight_0.5h | 0.241 | 0.539759 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.100 | 0.670489 |
| Re-illuminated_24h | Re-illuminated_24h | -0.064 | 0.703053 |