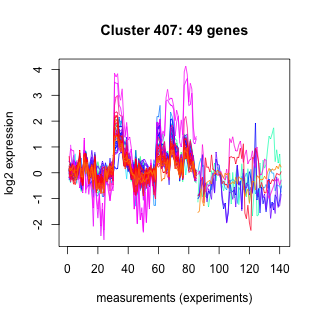
Phatr_hclust_0407 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015986 | ATP synthesis coupled proton transport | 0.0113395 | 1 | Phatr_hclust_0407 |
| GO:0006310 | DNA recombination | 0.152607 | 1 | Phatr_hclust_0407 |
| GO:0006281 | DNA repair | 0.141104 | 1 | Phatr_hclust_0407 |
| GO:0006260 | DNA replication | 0.109704 | 1 | Phatr_hclust_0407 |
| GO:0006633 | fatty acid biosynthesis | 0.0563567 | 1 | Phatr_hclust_0407 |
| GO:0051444 | negative regulation of ubiquitin-protein ligase activity | 0.00444554 | 1 | Phatr_hclust_0407 |
| GO:0051443 | positive regulation of ubiquitin-protein ligase activity | 0.00444554 | 1 | Phatr_hclust_0407 |
| GO:0006468 | protein amino acid phosphorylation | 0.542981 | 1 | Phatr_hclust_0407 |
| GO:0006464 | protein modification process | 0.00444554 | 1 | Phatr_hclust_0407 |
| GO:0016567 | protein ubiquitination | 0.276493 | 1 | Phatr_hclust_0407 |
|
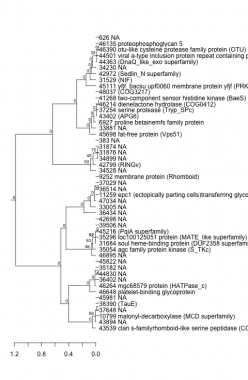
PHATRDRAFT_43539 : clan s-familyrhomboid-like serine peptidase (COG0705) |
PHATRDRAFT_46214 : dienelactone hydrolase (COG0412) |
||
|
PHATRDRAFT_35054 : agc family protein kinase (S_TKc) |
PHATRDRAFT_9252 : membrane protein (Rhomboid) |
PHATRDRAFT_41268 : two-component sensor histidine kinase (BaeS) |
|
|
PHATRDRAFT_10799 : malonyl-decarboxylase (MCD superfamily) |
PHATRDRAFT_31684 : soul heme-binding protein (DUF2358 superfamily) |
PHATRDRAFT_48037 : (COG3217) |
|
|
PHATRDRAFT_35296 : loc100125051 protein (MATE_like superfamily) |
PHATRDRAFT_42799 : (RINGv) |
PHATRDRAFT_45111 : yfjf_bacsu upf0060 membrane protein yfjf (PRK02237) |
|
|
PHATRDRAFT_38390 : (TauE) |
PHATRDRAFT_45216 : (PqiA superfamily) |
PHATRDRAFT_31529 : (NIF) |
|
|
PHATRDRAFT_42972 : (Sedlin_N superfamily) |
|||
|
PHATRDRAFT_46648 : platelet-binding glycoprotein |
|||
|
PHATRDRAFT_46264 : mgc68579 protein (HATPase_c) |
PHATRDRAFT_45698 : fat-free protein (Vps51) |
PHATRDRAFT_44363 : (DnaQ_like_exo superfamily) |
|
|
PHATRDRAFT_44501 : viral a-type inclusion protein repeat containing protein (SMC_prok_B) |
|||
|
PHATRDRAFT_6927 : proline betainemfs family protein |
PHATRDRAFT_46390 : otu-like cysteine protease family protein (OTU) |
||
|
PHATRDRAFT_11259 : epc1 (ectopically parting cells)transferring glycosyl groups (Glyco_transf_64 superfamily) |
PHATRDRAFT_43402 : (APG6) |
PHATRDRAFT_46135 : proteophosphoglycan 5 |
|
|
PHATRDRAFT_37254 : serine protease (Tryp_SPc) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_6h | Re-illuminated_6h | 0.705 | 0.000505051 |
| Copper_SH | Copper_SH | -0.612 | 0.000515464 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.205 | 0.02 |
| Silver_SH | Silver_SH | -0.272 | 0.000555556 |
| Cadmium_1.2mg | Cadmium_1.2mg | -0.008 | 0.925278 |
| BlueLight_24h | BlueLight_24h | 0.121 | 0.453606 |
| GreenLight_0.5h | GreenLight_0.5h | 1.282 | 0.000458716 |
| RedLight_6h | RedLight_6h | 0.628 | 0.000694444 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 1 |
| GreenLight_24h | GreenLight_24h | 0.206 | 0.0336842 |
| light_6hr | light_6hr | -0.193 | 0.217736 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.222 | 0.0981818 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.014 | 0.882066 |
| Ammonia_SH | Ammonia_SH | -0.430 | 0.0179545 |
| Simazine_SH | Simazine_SH | -0.336 | 0.000423729 |
| Oil_SH | Oil_SH | -0.171 | 0.0276163 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.461 | 0.000609756 |
| Cadmium_SH | Cadmium_SH | -0.472 | 0.000666667 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.290 | 0.00777778 |
| highlight_0to6h | highlight_0to6h | 0.128 | 0.2681 |
| BlueLight_0.5h | BlueLight_0.5h | 1.069 | 0.000462963 |
| Dispersant_SH | Dispersant_SH | -0.133 | 0.0908482 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.156 | 0.0630814 |
| light_15.5hr | light_15.5hr | -0.280 | 0.0860526 |
| light_10.5hr | light_10.5hr | -0.083 | 0.684336 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.054 | 0.49951 |
| Si_free | Si_free | 0.080 | 0.41213 |
| Re-illuminated_0.5h | Re-illuminated_0.5h | 1.376 | 0.000396825 |
| Mixture_SH | Mixture_SH | -0.534 | 0.00637255 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | 0.037 | 0.87365 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.204 | 0.221364 |
| BlueLight_6h | BlueLight_6h | 0.665 | 0.000581395 |
| highlight_12to48h | highlight_12to48h | -0.212 | 0.119383 |
| RedLight_24h | RedLight_24h | 0.411 | 0.000943396 |
| Dark_treated | Dark_treated | 1.554 | 0.000446429 |
| GreenLight_6h | GreenLight_6h | 0.679 | 0.000609756 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.015 | 0.673171 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.086 | 0.0975 |
| RedLight_0.5h | RedLight_0.5h | 1.118 | 0.000555556 |
| Green_vs_Red_6h | Green_vs_Red_6h | 0.051 | 0.793129 |
| Re-illuminated_24h | Re-illuminated_24h | 0.299 | 0.000657895 |