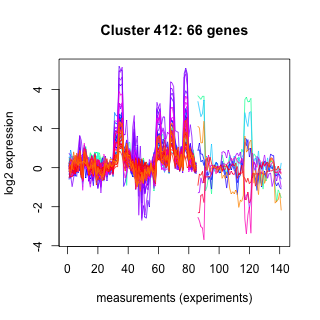
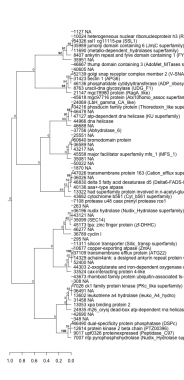
Phatr_hclust_0412 Hierarchical Clustering
Phaeodactylum tricornutum
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0009082 | branched chain family amino acid biosynthetic process | 0.00493949 | 1 | Phatr_hclust_0412 |
| GO:0006303 | double-strand break repair via nonhomologous end-joining | 0.0098558 | 1 | Phatr_hclust_0412 |
| GO:0006396 | RNA processing | 0.0304393 | 1 | Phatr_hclust_0412 |
| GO:0008654 | phospholipid biosynthesis | 0.0436294 | 1 | Phatr_hclust_0412 |
| GO:0006636 | fatty acid desaturation | 0.0483639 | 1 | Phatr_hclust_0412 |
| GO:0016192 | vesicle-mediated transport | 0.0624342 | 1 | Phatr_hclust_0412 |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.0624342 | 1 | Phatr_hclust_0412 |
| GO:0015992 | proton transport | 0.0670802 | 1 | Phatr_hclust_0412 |
| GO:0030001 | metal ion transport | 0.0763067 | 1 | Phatr_hclust_0412 |
| GO:0006470 | protein amino acid dephosphorylation | 0.0899841 | 1 | Phatr_hclust_0412 |
|
PHATRDRAFT_7007 : ntp pyrophosphohydrolase (Nudix_Hydrolase superfamily) |
PHATRDRAFT_10677 : copper-exporting atpase (ZntA) |
PHATRDRAFT_47026 : transmembrane protein 163 (Cation_efflux superfamily) |
PHATRDRAFT_45618 : mgc97716 protein (Atx10homo_assoc superfamily) |
|
PHATRDRAFT_9017 : upf0326 proteinexpressed (Peptidase_C97) |
PHATRDRAFT_11311 : silicon transporter (Silic_transp superfamily) |
PHATRDRAFT_21147 : mgc78980 protein (RagA_like) |
|
|
PHATRDRAFT_12614 : protein kinase 2 beta chain (PTZ00396) |
PHATRDRAFT_36789 : cyclin l |
PHATRDRAFT_8763 : uracil-dna glycosylase (UDG_F1) |
|
|
PHATRDRAFT_46490 : dual-specificity protein phosphatase (DSPc) |
PHATRDRAFT_45558 : major facilitator superfamily mfs_1 (MFS_1) |
PHATRDRAFT_46136 : phosphatidate cytidylyltransferase (ADP_ribosyl_GH) |
|
|
PHATRDRAFT_45173 : tpa: zinc finger protein (zf-DHHC) |
PHATRDRAFT_31423 : beclin 1 (APG6) |
||
|
PHATRDRAFT_24935 : rh26_orysj dead-box atp-dependent rna helicase 26 (DEXDc superfamily) |
PHATRDRAFT_35099 : (SEC14) |
PHATRDRAFT_52139 : golgi snap receptor complex member 2 (V-SNARE_C superfamily) |
|
|
PHATRDRAFT_13053 : xpa binding protein 2 |
|||
|
PHATRDRAFT_36196 : nudix hydrolase (Nudix_Hydrolase superfamily) |
PHATRDRAFT_37756 : (Abhydrolase_6) |
PHATRDRAFT_46667 : thump domain containing 3 (AdoMet_MTases superfamily) |
|
|
PHATRDRAFT_13602 : leukotriene a4 hydrolase (leuko_A4_hydro) |
PHATRDRAFT_7108 : protease u48 caax prenyl protease rce1 |
||
|
PHATRDRAFT_43682 : cytochrome b561 (Cyt_b561 superfamily) |
PHATRDRAFT_44966 : dna helicase |
PHATRDRAFT_8407 : ankyrin repeat and fyve domain containing 1 (FYVE) |
|
|
PHATRDRAFT_7026 : ck1 family protein kinase (PKc_like superfamily) |
PHATRDRAFT_13322 : had superfamily protein involved in n-acetyl-glucosamine catabolism-like (HAD-SF-IIA-hyp4) |
PHATRDRAFT_47127 : atp-dependent dna helicase (KU superfamily) |
PHATRDRAFT_11690 : (metallo-dependent_hydrolases superfamily) |
|
PHATRDRAFT_43673 : rhomboid family protein ubiquitin-associated ts-n domain-containing protein (Rhomboid superfamily) |
PHATRDRAFT_40136 : aaa+-type atpase |
PHATRDRAFT_35989 : jumonji domain containing 6 (JmjC superfamily) |
|
|
PHATRDRAFT_33524 : cax-interacting protein 4-like |
PHATRDRAFT_46830 : delta 5 fatty acid desaturase d5 (Delta6-FADS-like) |
PHATRDRAFT_34216 : phosducin family protein (Thioredoxin_like superfamily) |
PHATRDRAFT_54326 : ssl1 cg11115-pa (SSL1) |
|
PHATRDRAFT_44303 : 2-oxoglutarate and iron-dependent oxygenase domain containing 2 (2OG-FeII_Oxy_3 superfamily) |
PHATRDRAFT_24069 : (LbH_gamma_CA_like) |
PHATRDRAFT_10024 : heterogeneous nuclear ribonucleoprotein h3 (RRM_hnRNPH_ESRPs_RBM12_like) |
|
|
PHATRDRAFT_37109 : transmembrane efflux protein (ATG22) |
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| Re-illuminated_0.5h | Re-illuminated_0.5h | 2.182 | 0.000396825 |
| Simazine_SH | Simazine_SH | -0.621 | 0.000423729 |
| GreenLight_0.5h | GreenLight_0.5h | 1.717 | 0.000458716 |
| BlueLight_0.5h | BlueLight_0.5h | 2.048 | 0.000462963 |
| Re-illuminated_6h | Re-illuminated_6h | 0.571 | 0.000505051 |
| RedLight_0.5h | RedLight_0.5h | 1.332 | 0.000555556 |
| BlueLight_6h | BlueLight_6h | 0.507 | 0.000581395 |
| GreenLight_6h | GreenLight_6h | 0.572 | 0.000609756 |
| Re-illuminated_24h | Re-illuminated_24h | 0.398 | 0.000657895 |
| RedLight_6h | RedLight_6h | 0.629 | 0.000694444 |
| RedLight_24h | RedLight_24h | 0.386 | 0.000943396 |
| light_16hr_dark_4hr | light_16hr_dark_4hr | -0.234 | 0.000943396 |
| GreenLight_24h | GreenLight_24h | 0.284 | 0.000961538 |
| Dark_treated | Dark_treated | 1.104 | 0.00119048 |
| Blue_vs_Green_24h | Blue_vs_Green_24h | -0.130 | 0.00135135 |
| light_15.5hr | light_15.5hr | -0.358 | 0.00901639 |
| Ammonia_SH | Ammonia_SH | 0.395 | 0.0130332 |
| light_16hr_dark_30min | light_16hr_dark_30min | -0.329 | 0.0145299 |
| Blue_vs_Red_24h | Blue_vs_Red_24h | -0.232 | 0.0184211 |
| highlight_0to6h | highlight_0to6h | 0.179 | 0.0252212 |
| Salinity_15ppt_SH | Salinity_15ppt_SH | -0.141 | 0.0595238 |
| Mixture_SH | Mixture_SH | 0.323 | 0.0659341 |
| Dispersed_oil_SH | Dispersed_oil_SH | -0.107 | 0.123985 |
| Silver_SH | Silver_SH | 0.096 | 0.131145 |
| BlueLight_24h | BlueLight_24h | 0.154 | 0.135294 |
| Cadmium_SH | Cadmium_SH | -0.108 | 0.14835 |
| light_10.5hr | light_10.5hr | -0.217 | 0.150209 |
| Green_vs_Red_24h | Green_vs_Red_24h | -0.102 | 0.191566 |
| Oil_SH | Oil_SH | -0.079 | 0.192429 |
| dark_8hr_light_3hr | dark_8hr_light_3hr | -0.151 | 0.200584 |
| Blue_vs_Green_6h | Blue_vs_Green_6h | -0.066 | 0.255556 |
| Blue_vs_Red_6h | Blue_vs_Red_6h | -0.122 | 0.331818 |
| Cadmium_1.2mg | Cadmium_1.2mg | 0.042 | 0.373377 |
| Si_free | Si_free | 0.068 | 0.4485 |
| light_6hr | light_6hr | -0.101 | 0.48137 |
| Green_vs_Red_6h | Green_vs_Red_6h | -0.057 | 0.730161 |
| Dispersant_SH | Dispersant_SH | 0.011 | 0.898712 |
| Cadmium_0.12mg | Cadmium_0.12mg | -0.005 | 0.911229 |
| highlight_12to48h | highlight_12to48h | -0.013 | 0.935227 |
| Copper_SH | Copper_SH | 0.005 | 0.948265 |
| light_16hr_dark_7.5hr | light_16hr_dark_7.5hr | 0.000 | 0.980594 |