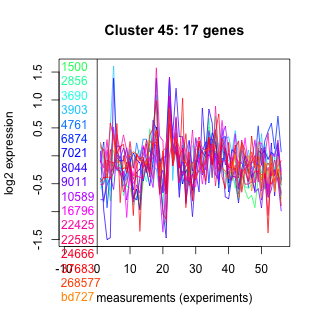
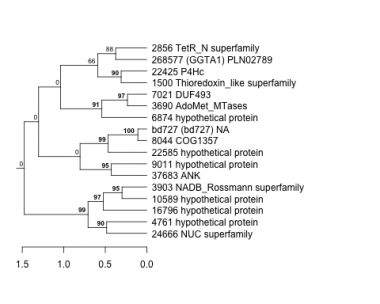
Thaps_hclust_0045 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015986 | ATP synthesis coupled proton transport | 0.0466732 | 1 | Thaps_hclust_0045 |
| GO:0006118 | electron transport | 0.24643 | 1 | Thaps_hclust_0045 |
| GO:0008152 | metabolism | 0.355748 | 1 | Thaps_hclust_0045 |
| GO:0018346 | protein amino acid prenylation | 0.00515995 | 1 | Thaps_hclust_0045 |
| GO:0019538 | protein metabolism | 0.0466732 | 1 | Thaps_hclust_0045 |
|
1500 : Thioredoxin_like superfamily |
6874 : hypothetical protein |
10589 : hypothetical protein |
24666 : NUC superfamily |
|
2856 : TetR_N superfamily |
7021 : DUF493 |
16796 : hypothetical protein |
37683 : ANK |
|
3690 : AdoMet_MTases |
8044 : COG1357 |
22425 : P4Hc |
268577 : (GGTA1) PLN02789 |
|
3903 : NADB_Rossmann superfamily |
9011 : hypothetical protein |
22585 : hypothetical protein |
bd727 : (bd727) NA |
|
4761 : hypothetical protein |
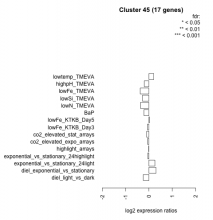
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | -0.243 | 0.301 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.058 | 0.859 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.024 | 0.923 |
| BaP | BaP | -0.214 | 0.386 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.046 | 0.729 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.095 | 0.732 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.210 | 0.451 |
| highpH_TMEVA | highpH_TMEVA | -0.198 | 0.243 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.056 | 0.903 |
| lowFe_TMEVA | lowFe_TMEVA | -0.374 | 0.215 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.258 | 0.482 |
| lowN_TMEVA | lowN_TMEVA | -0.362 | 0.31 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.305 | 0.11 |
| lowSi_TMEVA | lowSi_TMEVA | -0.275 | 0.761 |
| highlight_arrays | highlight_arrays | -0.010 | 0.965 |