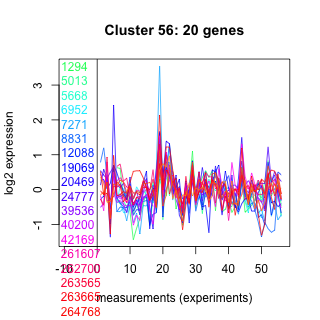
Thaps_hclust_0056 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006350 | transcription | 0.0000176 | 0.0641837 | Thaps_hclust_0056 |
| GO:0006354 | RNA elongation | 0.00742418 | 1 | Thaps_hclust_0056 |
| GO:0006413 | translational initiation | 0.0438152 | 1 | Thaps_hclust_0056 |
| GO:0006412 | protein biosynthesis | 0.269889 | 1 | Thaps_hclust_0056 |
| GO:0006118 | electron transport | 0.399203 | 1 | Thaps_hclust_0056 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.446038 | 1 | Thaps_hclust_0056 |
| GO:0008152 | metabolism | 0.546944 | 1 | Thaps_hclust_0056 |
|
1294 : hypothetical protein |
8831 : UAA |
39536 : Zn-ribbon_RPB9 |
263565 : RNAP_I_III_AC19 |
|
5013 : hypothetical protein |
12088 : DRG |
40200 : TRX_reduct |
263665 : (RT17) rpsQ |
|
5668 : hypothetical protein |
19069 : hypothetical protein |
42169 : Nop10p |
264768 : Tim17 superfamily |
|
6952 : hypothetical protein |
20469 : PLN00032 |
261607 : IF4E |
270037 : (Tp_CCCH5) RRM_U2AF35B |
|
7271 : hypothetical protein |
24777 : WGG superfamily |
262700 : Dpy-30 superfamily |
270256 : hypothetical protein |
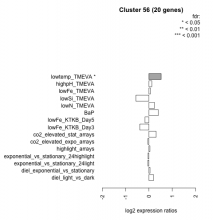
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| BaP | BaP | 0.401 | 0.0602 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | -0.047 | 0.926 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.309 | 0.169 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.049 | 0.821 |
| diel_light_vs_dark | diel_light_vs_dark | 0.199 | 0.379 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | -0.041 | 0.74 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | -0.046 | 0.902 |
| highlight_arrays | highlight_arrays | 0.045 | 0.808 |
| highpH_TMEVA | highpH_TMEVA | 0.126 | 0.466 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | -0.390 | 0.0893 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.169 | 0.42 |
| lowFe_TMEVA | lowFe_TMEVA | 0.070 | 0.857 |
| lowN_TMEVA | lowN_TMEVA | 0.242 | 0.505 |
| lowSi_TMEVA | lowSi_TMEVA | -0.559 | 0.22 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.525 | 0.0421 |