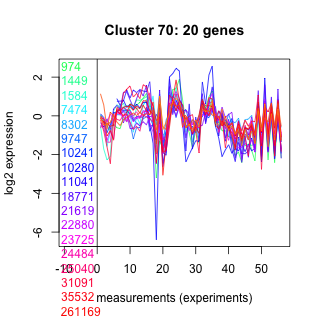
Thaps_hclust_0070 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0015969 | guanosine tetraphosphate metabolic process | 0.00330783 | 1 | Thaps_hclust_0070 |
| GO:0006012 | galactose metabolism | 0.0196937 | 1 | Thaps_hclust_0070 |
| GO:0008152 | metabolism | 0.755516 | 1 | Thaps_hclust_0070 |
| GO:0006241 | CTP biosynthesis | 0.0229404 | 1 | Thaps_hclust_0070 |
| GO:0006183 | GTP biosynthesis | 0.0229404 | 1 | Thaps_hclust_0070 |
| GO:0006228 | UTP biosynthesis | 0.0229404 | 1 | Thaps_hclust_0070 |
| GO:0009225 | nucleotide-sugar metabolism | 0.0294036 | 1 | Thaps_hclust_0070 |
| GO:0007155 | cell adhesion | 0.0948904 | 1 | Thaps_hclust_0070 |
| GO:0006260 | DNA replication | 0.10986 | 1 | Thaps_hclust_0070 |
| GO:0006118 | electron transport | 0.218688 | 1 | Thaps_hclust_0070 |
|
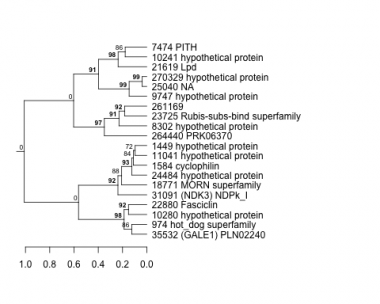
974 : hot_dog superfamily |
9747 : hypothetical protein |
21619 : Lpd |
31091 : (NDK3) NDPk_I |
|
1449 : hypothetical protein |
10241 : hypothetical protein |
22880 : Fasciclin |
35532 : (GALE1) PLN02240 |
|
1584 : cyclophilin |
10280 : hypothetical protein |
23725 : Rubis-subs-bind superfamily |
261169 : |
|
7474 : PITH |
11041 : hypothetical protein |
24484 : hypothetical protein |
264440 : PRK06370 |
|
8302 : hypothetical protein |
18771 : MORN superfamily |
25040 : NA |
270329 : hypothetical protein |
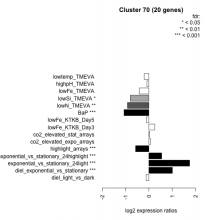
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| BaP | BaP | -1.070 | 0.00037 |
| highlight_arrays | highlight_arrays | -0.572 | 0.000442 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.547 | 0.000526 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 1.750 | 0.000581 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 1.010 | 0.000602 |
| lowN_TMEVA | lowN_TMEVA | -0.919 | 0.00313 |
| lowSi_TMEVA | lowSi_TMEVA | -0.797 | 0.0367 |
| lowFe_TMEVA | lowFe_TMEVA | -0.412 | 0.108 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.252 | 0.296 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.199 | 0.441 |
| diel_light_vs_dark | diel_light_vs_dark | -0.120 | 0.636 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.104 | 0.637 |
| highpH_TMEVA | highpH_TMEVA | -0.086 | 0.644 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.070 | 0.833 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.034 | 0.91 |