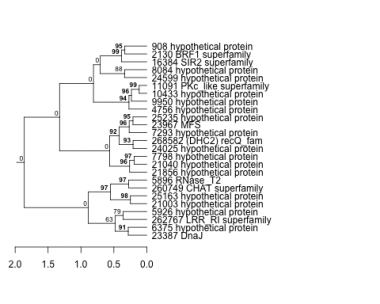
Thaps_hclust_0084 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006342 | chromatin silencing | 0.0129601 | 1 | Thaps_hclust_0084 |
| GO:0006259 | DNA metabolism | 0.0200978 | 1 | Thaps_hclust_0084 |
| GO:0006306 | DNA methylation | 0.0412459 | 1 | Thaps_hclust_0084 |
| GO:0006355 | regulation of transcription, DNA-dependent | 0.0681942 | 1 | Thaps_hclust_0084 |
| GO:0006457 | protein folding | 0.173725 | 1 | Thaps_hclust_0084 |
| GO:0006468 | protein amino acid phosphorylation | 0.270534 | 1 | Thaps_hclust_0084 |
|
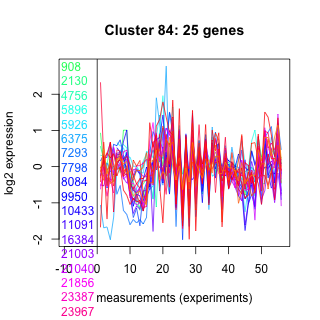
908 : hypothetical protein |
7798 : hypothetical protein |
21003 : hypothetical protein |
24599 : hypothetical protein |
|
2130 : BRF1 superfamily |
8084 : hypothetical protein |
21040 : hypothetical protein |
25163 : hypothetical protein |
|
4756 : hypothetical protein |
9950 : hypothetical protein |
21856 : hypothetical protein |
25235 : hypothetical protein |
|
5896 : RNase_T2 |
10433 : hypothetical protein |
23387 : DnaJ |
260749 : CHAT superfamily |
|
5926 : hypothetical protein |
11091 : PKc_like superfamily |
23967 : MFS |
262767 : LRR_RI superfamily |
|
6375 : hypothetical protein |
16384 : SIR2 superfamily |
24025 : hypothetical protein |
268582 : (DHC2) recQ_fam |
|
7293 : hypothetical protein |
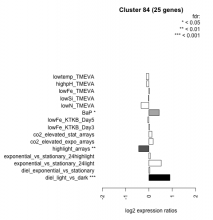
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.903 | 0.000485 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.027 | 0.926 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | -0.048 | 0.82 |
| BaP | BaP | 0.443 | 0.0175 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.058 | 0.579 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | 0.132 | 0.599 |
| lowtemp_TMEVA | lowtemp_TMEVA | -0.117 | 0.645 |
| highpH_TMEVA | highpH_TMEVA | -0.103 | 0.51 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.182 | 0.213 |
| lowFe_TMEVA | lowFe_TMEVA | 0.018 | 0.975 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.527 | 0.0583 |
| lowN_TMEVA | lowN_TMEVA | -0.335 | 0.293 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.042 | 0.844 |
| lowSi_TMEVA | lowSi_TMEVA | -0.017 | 1 |
| highlight_arrays | highlight_arrays | -0.437 | 0.00157 |