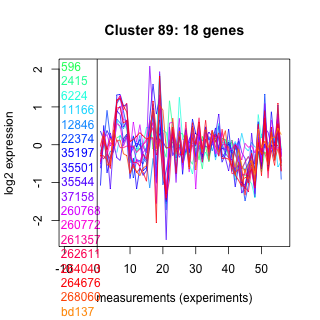
Thaps_hclust_0089 Hierarchical Clustering
Thalassiosira pseudonana
| GO ID | Go Term | p-value | q-value | Cluster |
|---|---|---|---|---|
| GO:0006084 | acetyl-CoA metabolism | 0.000826959 | 1 | Thaps_hclust_0089 |
| GO:0017004 | cytochrome complex assembly | 0.000826959 | 1 | Thaps_hclust_0089 |
| GO:0043087 | regulation of GTPase activity | 0.00824653 | 1 | Thaps_hclust_0089 |
| GO:0006468 | protein amino acid phosphorylation | 0.164893 | 1 | Thaps_hclust_0089 |
|
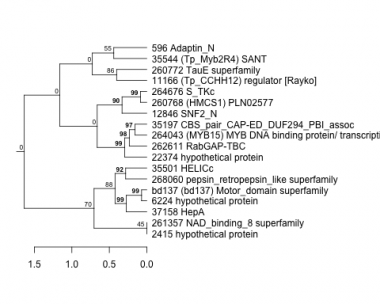
596 : Adaptin_N |
22374 : hypothetical protein |
260768 : (HMCS1) PLN02577 |
264043 : (MYB15) MYB DNA binding protein/ transcription factor-like protein |
|
2415 : hypothetical protein |
35197 : CBS_pair_CAP-ED_DUF294_PBI_assoc |
260772 : TauE superfamily |
264676 : S_TKc |
|
6224 : hypothetical protein |
35501 : HELICc |
261357 : NAD_binding_8 superfamily |
268060 : pepsin_retropepsin_like superfamily |
|
11166 : (Tp_CCHH12) regulator [Rayko] |
35544 : (Tp_Myb2R4) SANT |
262611 : RabGAP-TBC |
bd137 : (bd137) Motor_domain superfamily |
|
12846 : SNF2_N |
37158 : HepA |
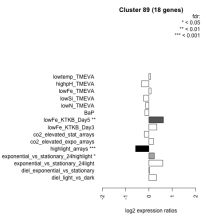
| Condition | Condition | Difference | FDR |
|---|---|---|---|
| diel_light_vs_dark | diel_light_vs_dark | 0.321 | 0.136 |
| lowFe_KTKB_Day3 | lowFe_KTKB_Day3 | 0.339 | 0.165 |
| lowFe_KTKB_Day5 | lowFe_KTKB_Day5 | 0.622 | 0.00244 |
| BaP | BaP | -0.174 | 0.483 |
| exponential_vs_stationary_24highlight | exponential_vs_stationary_24highlight | 0.233 | 0.0349 |
| co2_elevated_stat_arrays | co2_elevated_stat_arrays | -0.195 | 0.444 |
| lowtemp_TMEVA | lowtemp_TMEVA | 0.068 | 0.825 |
| highpH_TMEVA | highpH_TMEVA | -0.321 | 0.0533 |
| co2_elevated_expo_arrays | co2_elevated_expo_arrays | 0.201 | 0.224 |
| lowFe_TMEVA | lowFe_TMEVA | 0.095 | 0.813 |
| exponential_vs_stationary_24light | exponential_vs_stationary_24light | 0.596 | 0.0648 |
| lowN_TMEVA | lowN_TMEVA | -0.133 | 0.741 |
| diel_exponential_vs_stationary | diel_exponential_vs_stationary | 0.033 | 0.896 |
| lowSi_TMEVA | lowSi_TMEVA | -0.227 | 0.838 |
| highlight_arrays | highlight_arrays | -0.573 | 0.000442 |